Using colour and annotations for effective story telling
Workshops for Ukraine | #rstats #ggplot | 20th April 2023
Hi there 👋 !
👩 Cara Thompson
👩💻 A love for patterns |> Psychology PhD |> Analysis of postgraduate medical examinations |> Data Visualisation Consultant
💙 Helping others maximise the impact of their expertise
Find out more: cararthompson.com/about
Today’s goal
To equip you with some design tips and coding tricks to enhance the storytelling capabilities of your plots.
- Explore how to be less dependent on annotations by using intuitive colour palettes
- Illustrate ways in which we can use text colour and fonts to add text hierarchy
- Add in story-enhancing annotations and data highlights to draw attention to the key data patterns
- Package up bits of reusable
Rcode - Introduce you to
{ggtext},{geomtextpath}and{gghighlight} - Q&A
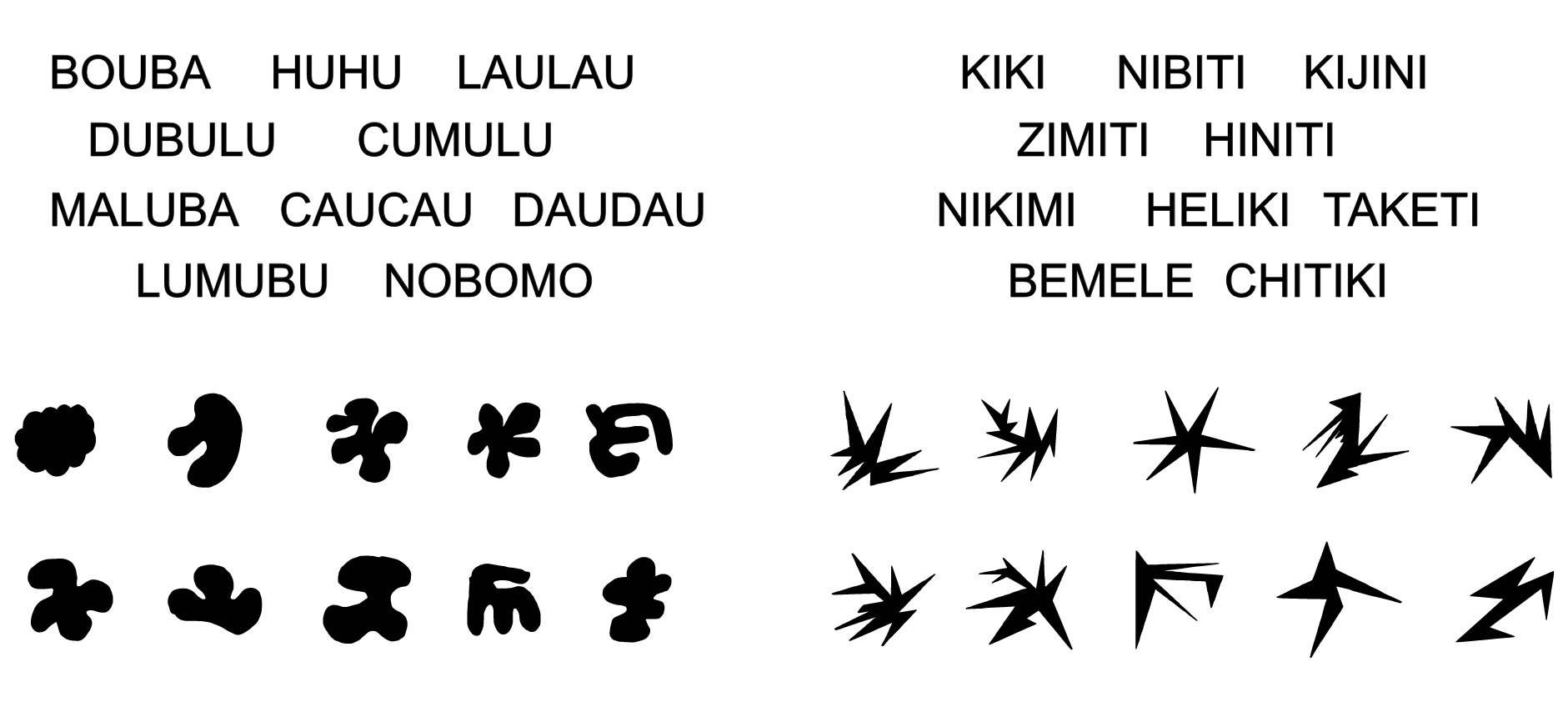
“Intuitive colour palettes?”
Find out more: en.wikipedia.org/wiki/Bouba/kiki_effect
“Intuitive colour palettes?”
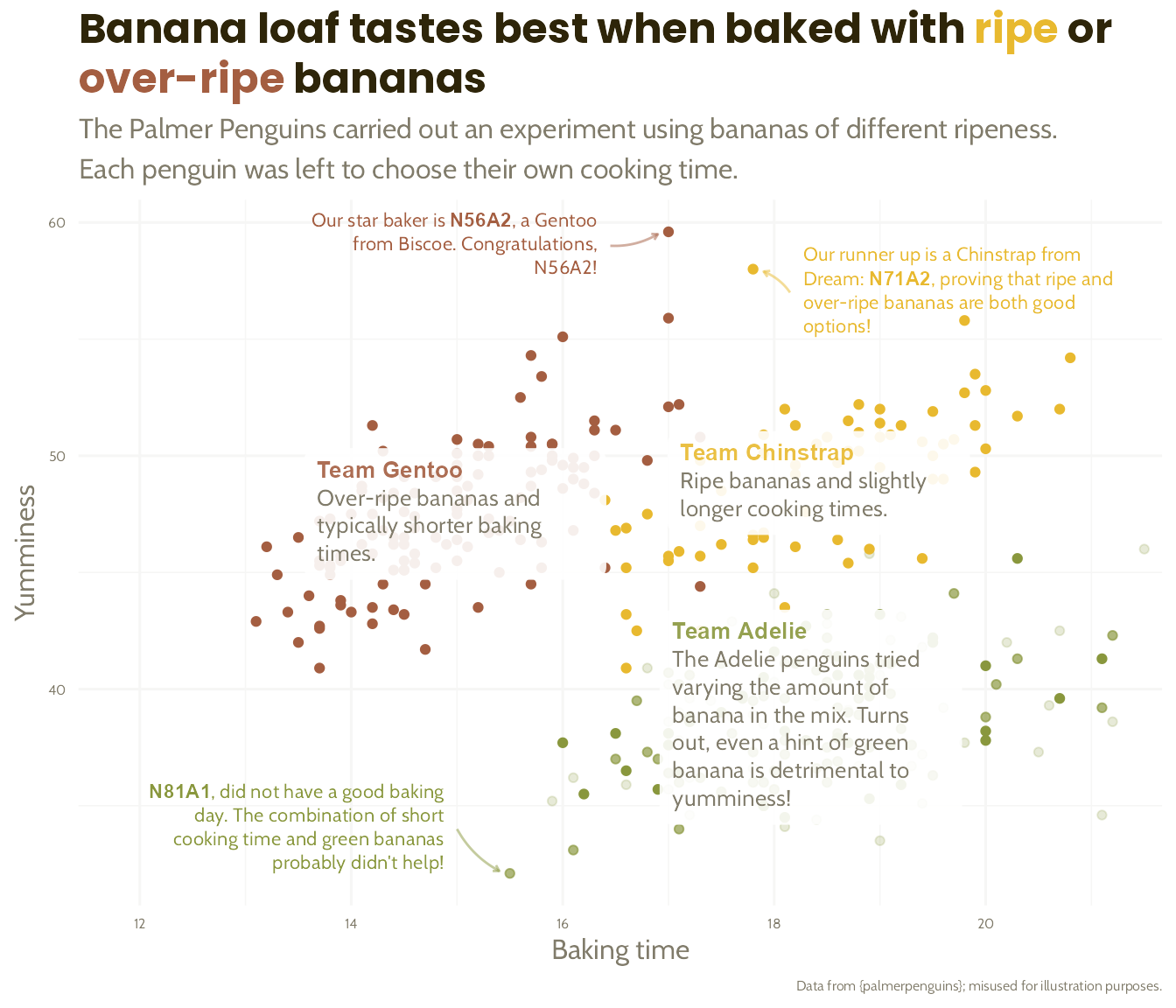
The Great Penguin Bake Off
The penguins had a baking competition to see which species could make the best banana loaf. Each species was given bananas of a different level of ripeness.
The Great Penguin Bake Off
The penguins had a baking competition to see which species could make the best banana loaf. Each species was given bananas of a different level of ripeness.
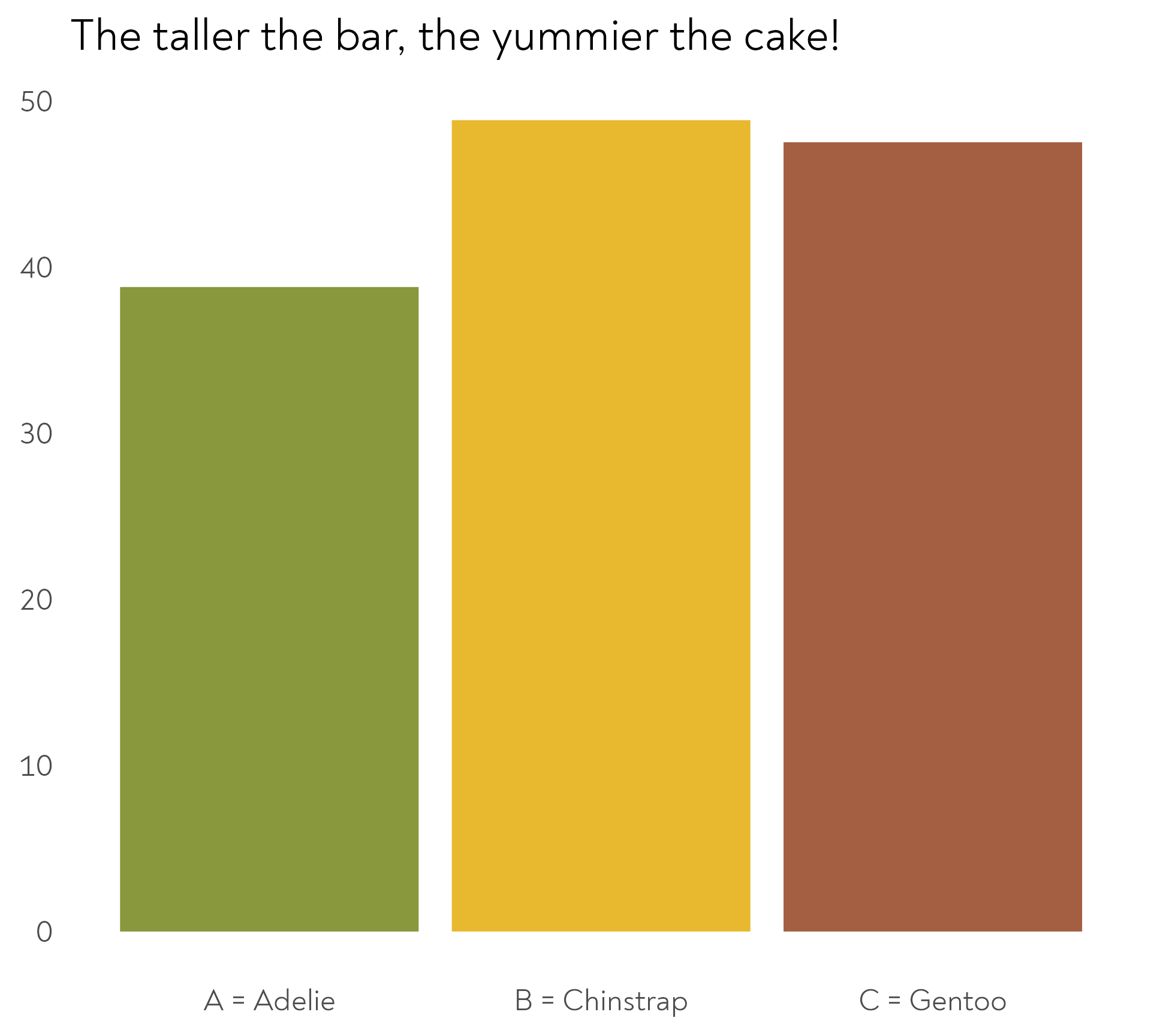
The Great Penguin Bake Off
The Adelie penguins decided to experiment with different quantities of banana in their mix. Each island chose a different quantity.

The Great Penguin Bake Off
The Adelie penguins decided to experiment with different quantities of unripe banana in their mix. Each island chose a different quantity.

The Great Penguin Bake Off
They decided to go on a retreat to plan their bakes in different locations
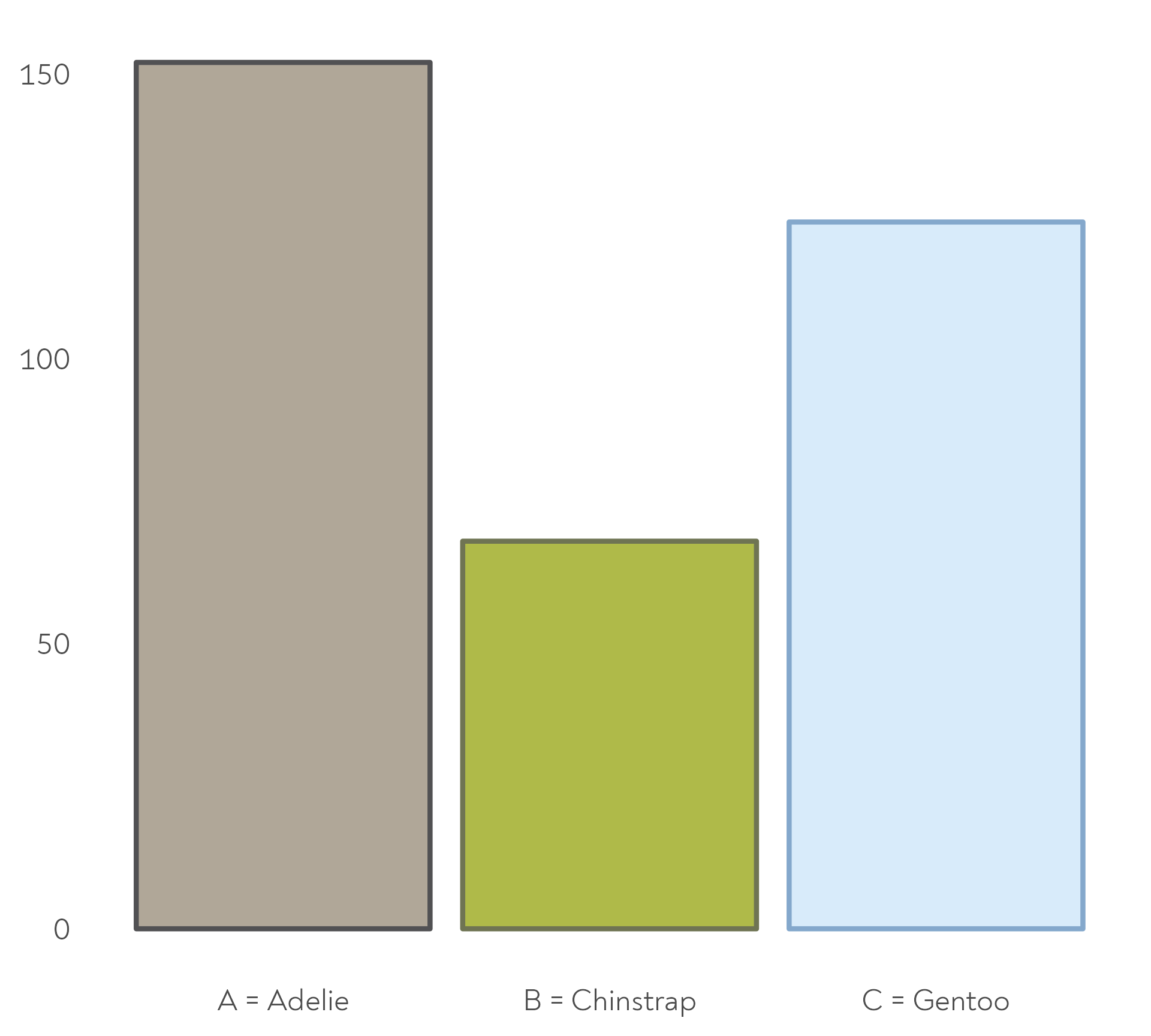
The Great Penguin Bake Off
Each species was allowed to invite a different mentor…
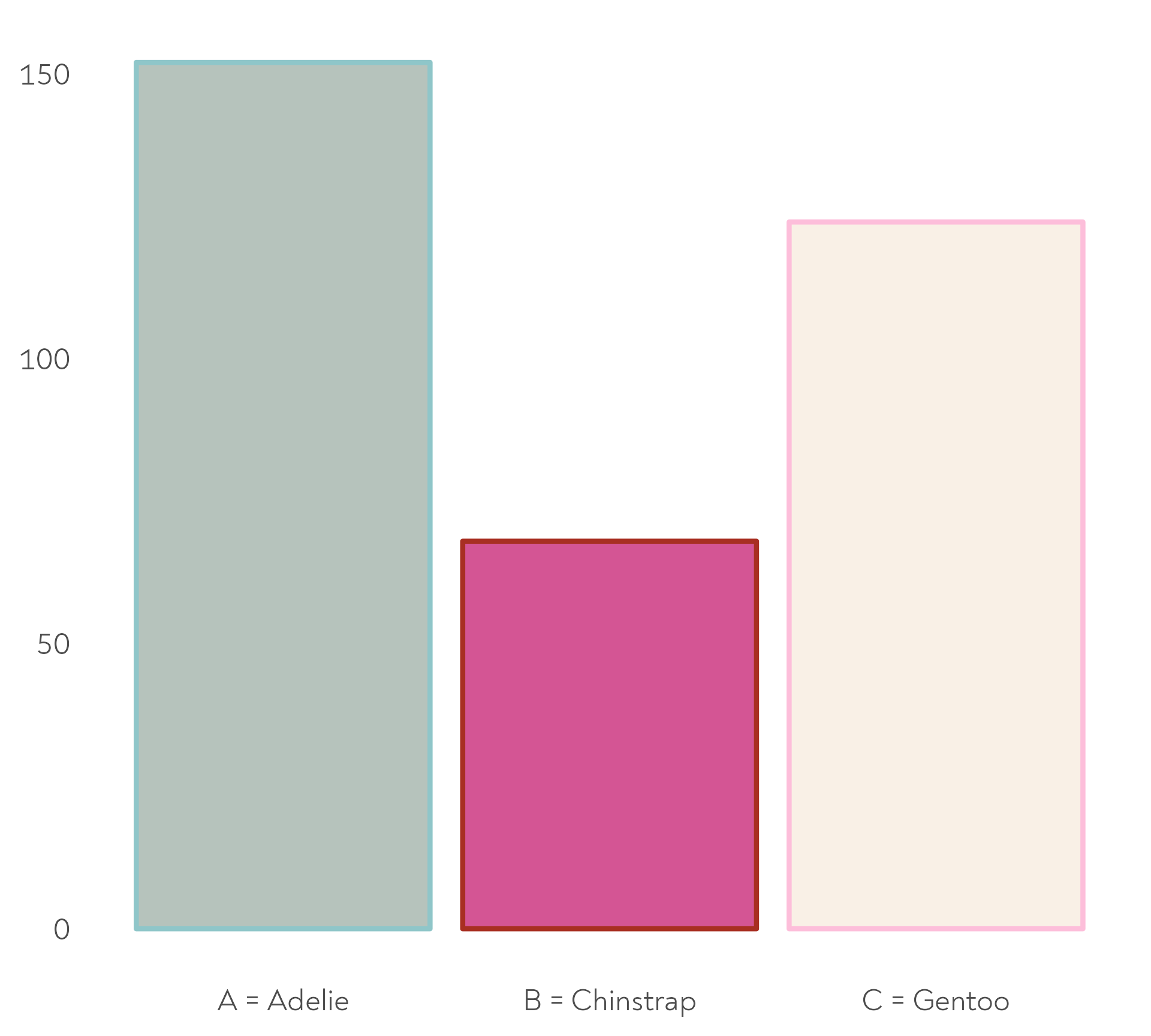
The Great Penguin Bake Off
… and to choose a type of snack between practice bakes
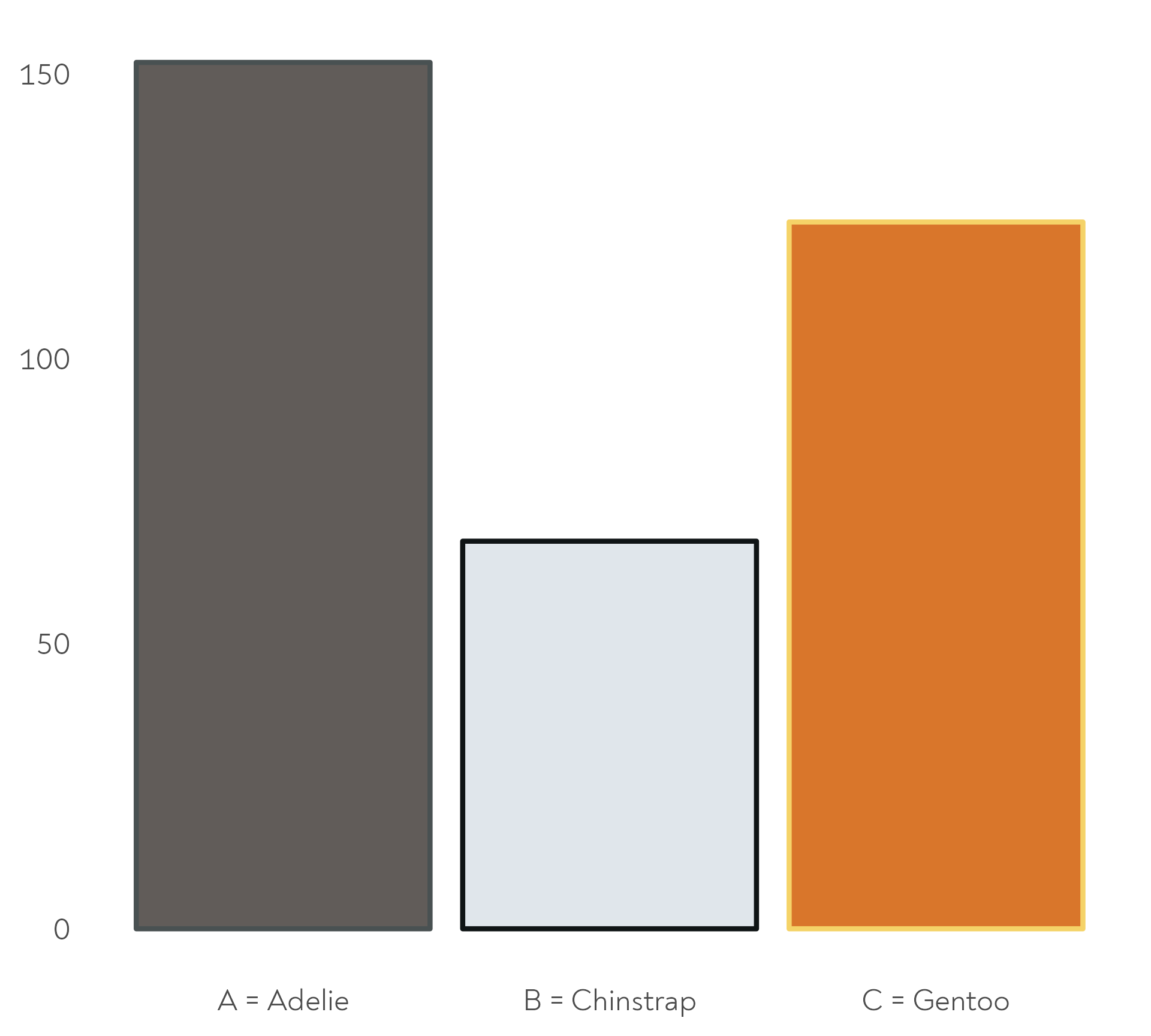
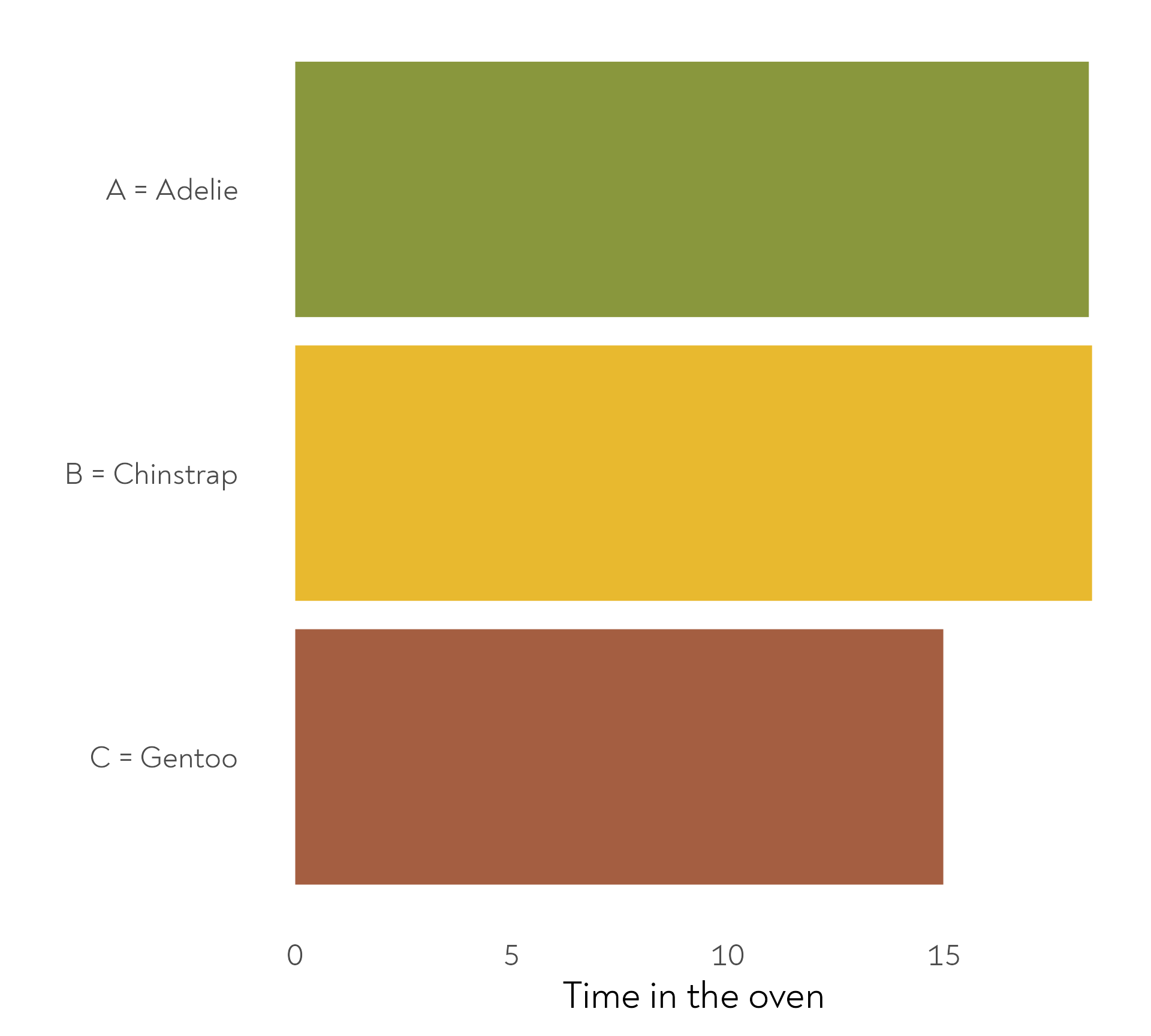
The Great Penguin Bake Off - Bonus round!
The penguins also baked their cakes for different amounts of time. Here are the mean durations per species.
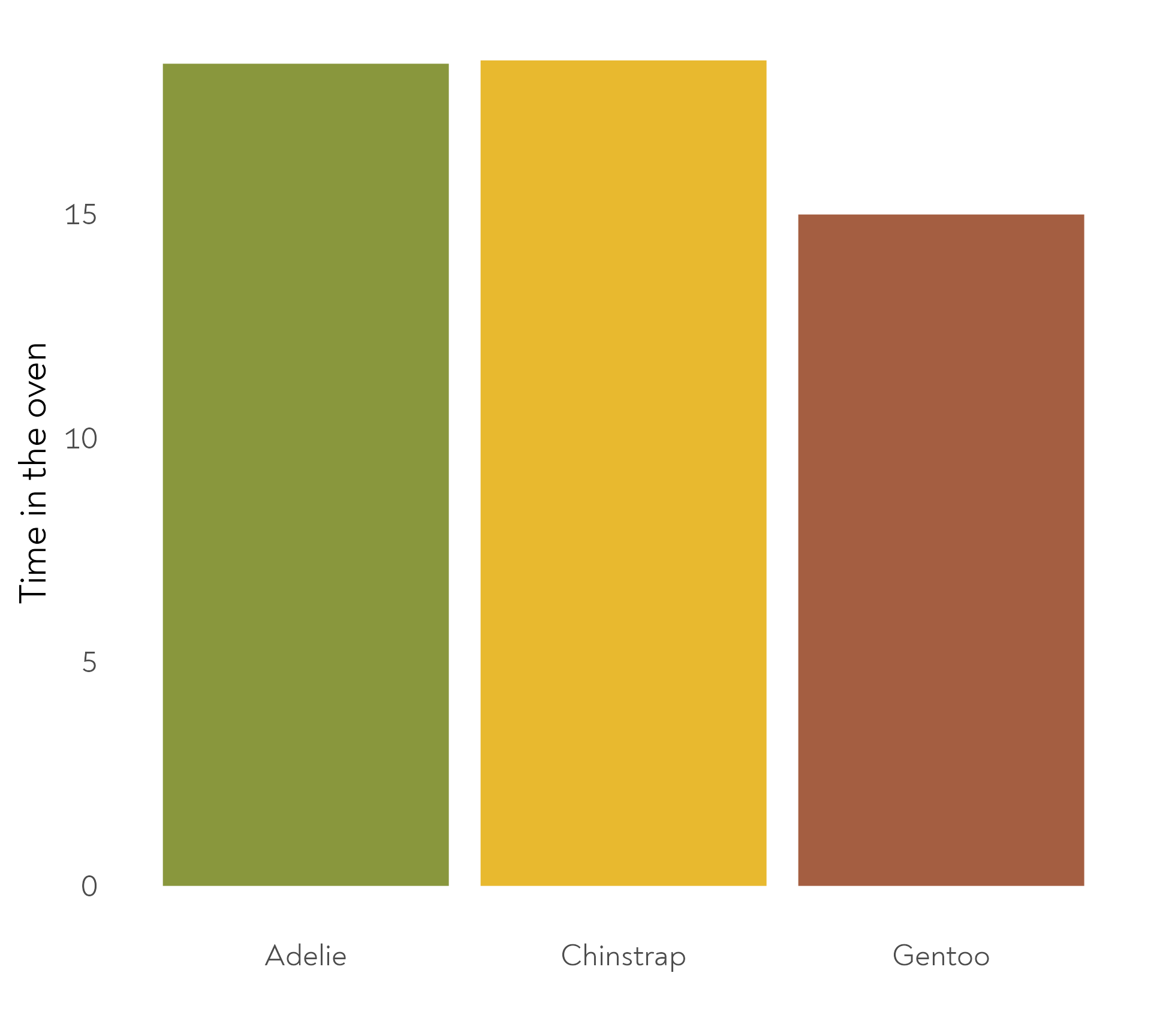
The Great Penguin Bake Off - Bonus round!
The penguins also baked their cakes for different amounts of time. Here are the mean durations per species.

Five tips for storytelling
#1 - Use colour (and orientation) purposefully
#2 - Add text hierarchy
#3 - Reduce unnecessary eye movement
#4 - Highlight important patterns
#5 - See how much you can declutter
Bonus track - {gghighlight}, Tee pipe and curved arrows
#1 Use colour purposefully
All about making it easier to remember what is what
Picking colours is hard! Let others help you!
- Your brand guidelines
- A photo + something like imagecolorpicker.com to pick out colours
#1 Use colour purposefully

#1 Use colour purposefully
#1 Use colour purposefully
#1 Use colour purposefully
- Your department brand guidelines
- A photo + something like imagecolorpicker.com to pick out colours
- Take inspiration from other dataviz / art you like
#1 Use colour purposefully

#1 Use colour purposefully
- Your department brand guidelines
- A photo + something like imagecolorpicker.com to pick out colours
- Take inspiration from other dataviz / art you like
- Google images and “[whatever you like] palette”
#1 Use colour purposefully

#1 Use colour purposefully
- Your department brand guidelines
- A photo + something like imagecolorpicker.com to pick out colours
- Take inspiration from other dataviz / art you like
- Google images and “[whatever you like] palette”
- Or… start from the colour wheel and read around how best to use it
- Using a tool like paletton.com makes it easier!
Find out more: blog.datawrapper.de/colors-for-data-vis-style-guides/
#1 Use colour purposefully
Quick tip: Viewing your colours
#1 Use colour purposefully
Quick tip: Naming and viewing your colours
#1 Use colour purposefully
A few things to bear in mind
- Accessibility
colorblindr::cvd_grid()
#1 Use colour purposefully
A few things to bear in mind
- Accessibility
colorblindr::cvd_grid()- colourcontrast.cc
#1 Use colour purposefully
A few things to bear in mind
- Accessibility
- Race/Ethnicity - avoid stereotypes and be mindful of unintended messages
- Colour intensity - “more is more”
#1 Use colour purposefully
A few things to bear in mind
- Accessibility
- Race/Ethnicity - avoid stereotypes and be mindful of unintended messages
- Colour intensity - “more is more”
#1 Use colour purposefully
A few things to bear in mind
- Accessibility
- Race/Ethnicity - avoid stereotypes and be mindful of unintended messages
- Colour intensity - “more is more” - consider “dark mode”

#1 Use colour purposefully
A few things to bear in mind
- Accessibility
- Race/Ethnicity - avoid stereotypes and be mindful of unintended messages
- Colour intensity - “more is more” - consider “dark mode”
Over to you!
- Think about the concepts in your own data
- Look for images related to those concepts
- Extract the colour codes you need -
imagecolorpicker.com - Check what they look like together -
monochromeR::view_palette() - Assess using
colorblindr::cvd_grid()
See you in 5 minutes! 📊 🎨 ☕
05:00
Let’s get coding!
Setting up our first plot
Using the ToothGrowth dataset
Setting up our first plot
With a few tips along the way
Setting up our first plot
With a few tips along the way
Setting up our first plot
With a few tips along the way
Setting up our first plot
Mini tip: get rid of abbreviations
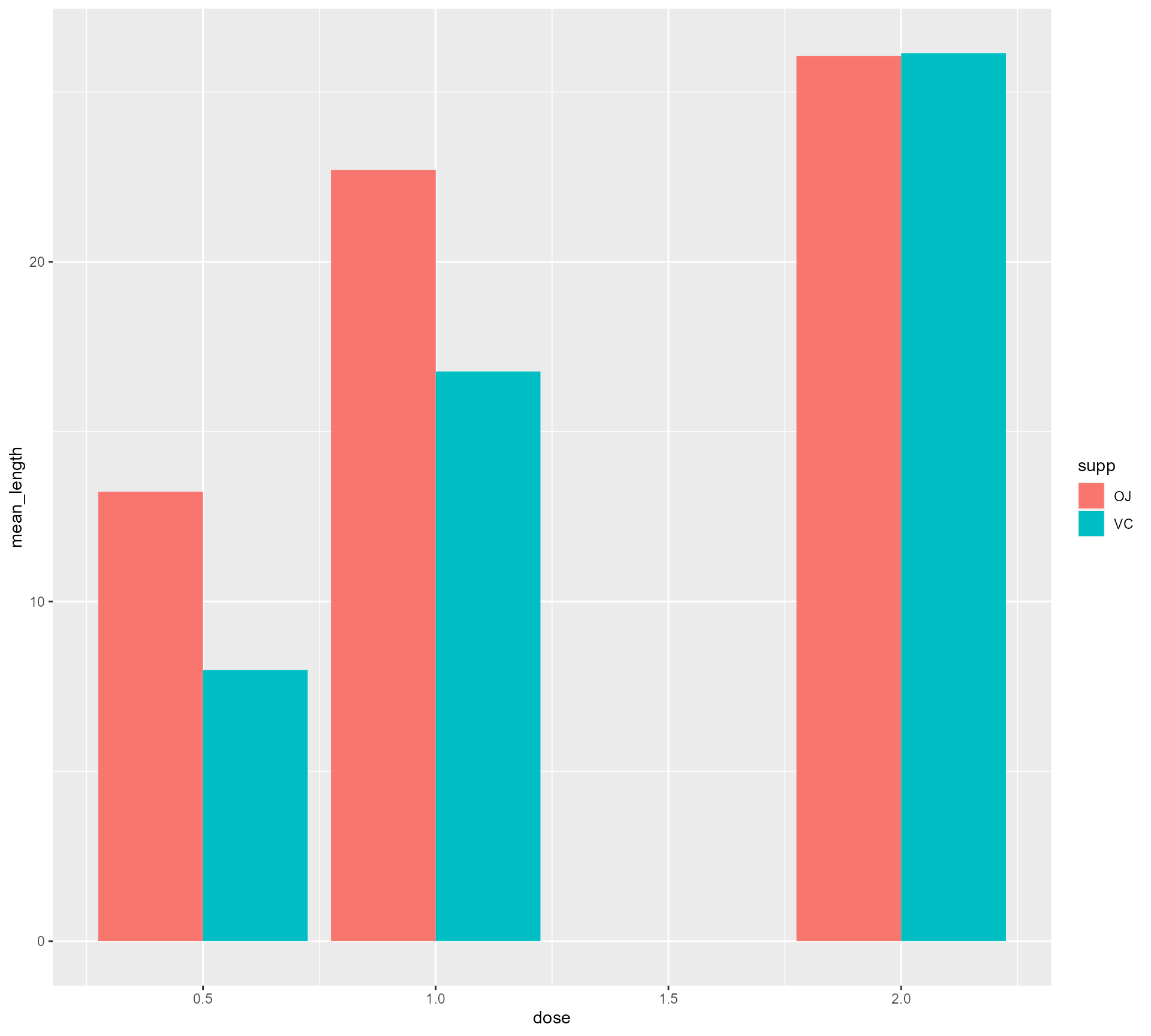
ToothGrowth %>%
mutate(supplement =
case_when(supp == "OJ" ~ "Orange Juice",
supp == "VC" ~ "Vitamin C",
TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
ggplot(aes(x = dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
position = "dodge",
colour = "#FFFFFF",
size = 2)Setting up our first plot
Mini tip: theme_minimal()
ToothGrowth %>%
mutate(supplement =
case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
ggplot(aes(x = dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
position = "dodge",
colour = "#FFFFFF",
size = 2) +
theme_minimal()Setting up our first plot
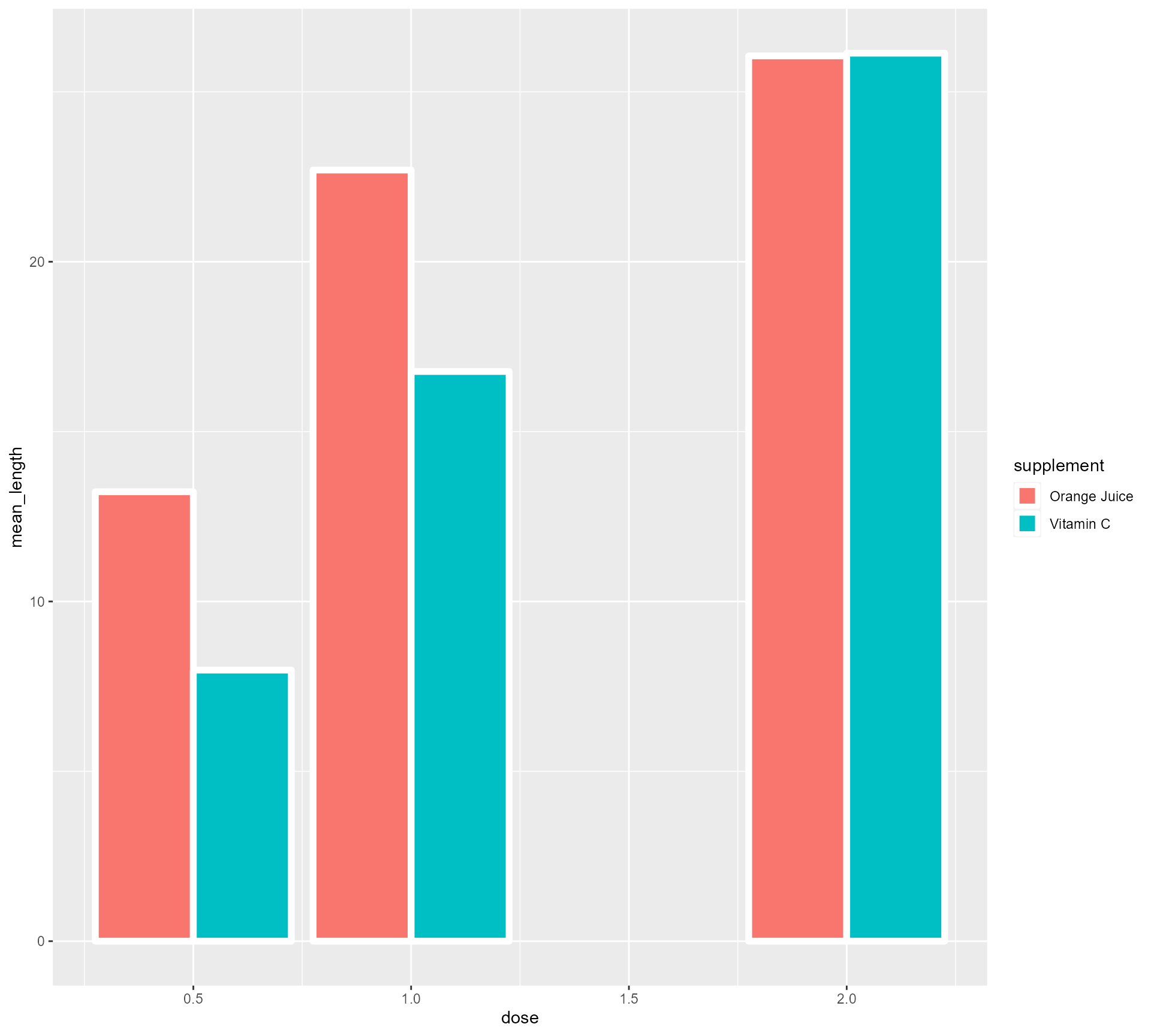
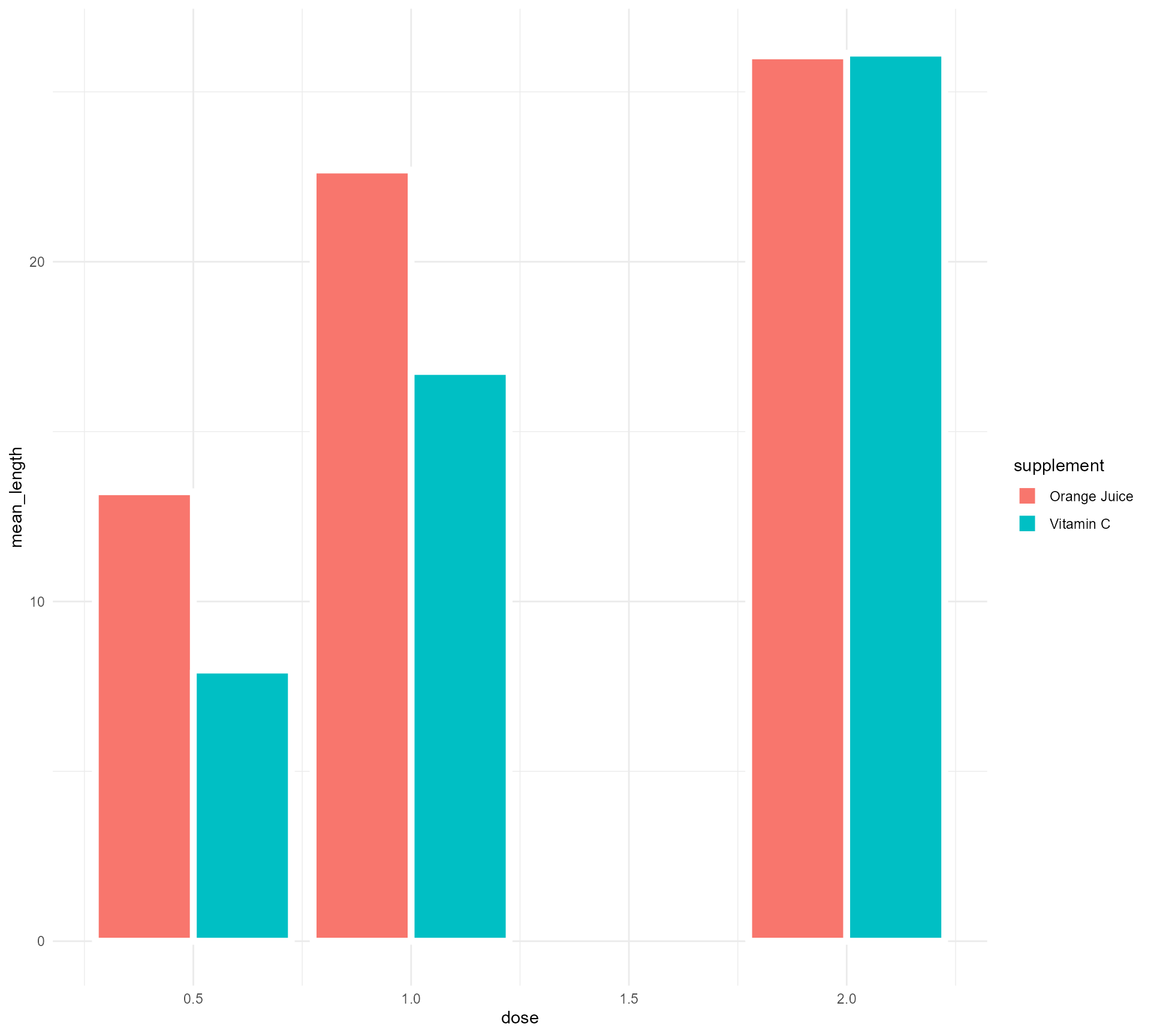
Turning Dose into a categorical variable (fear not!)
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
position = "dodge",
colour = "#FFFFFF",
size = 2) +
theme_minimal()Setting up our first plot
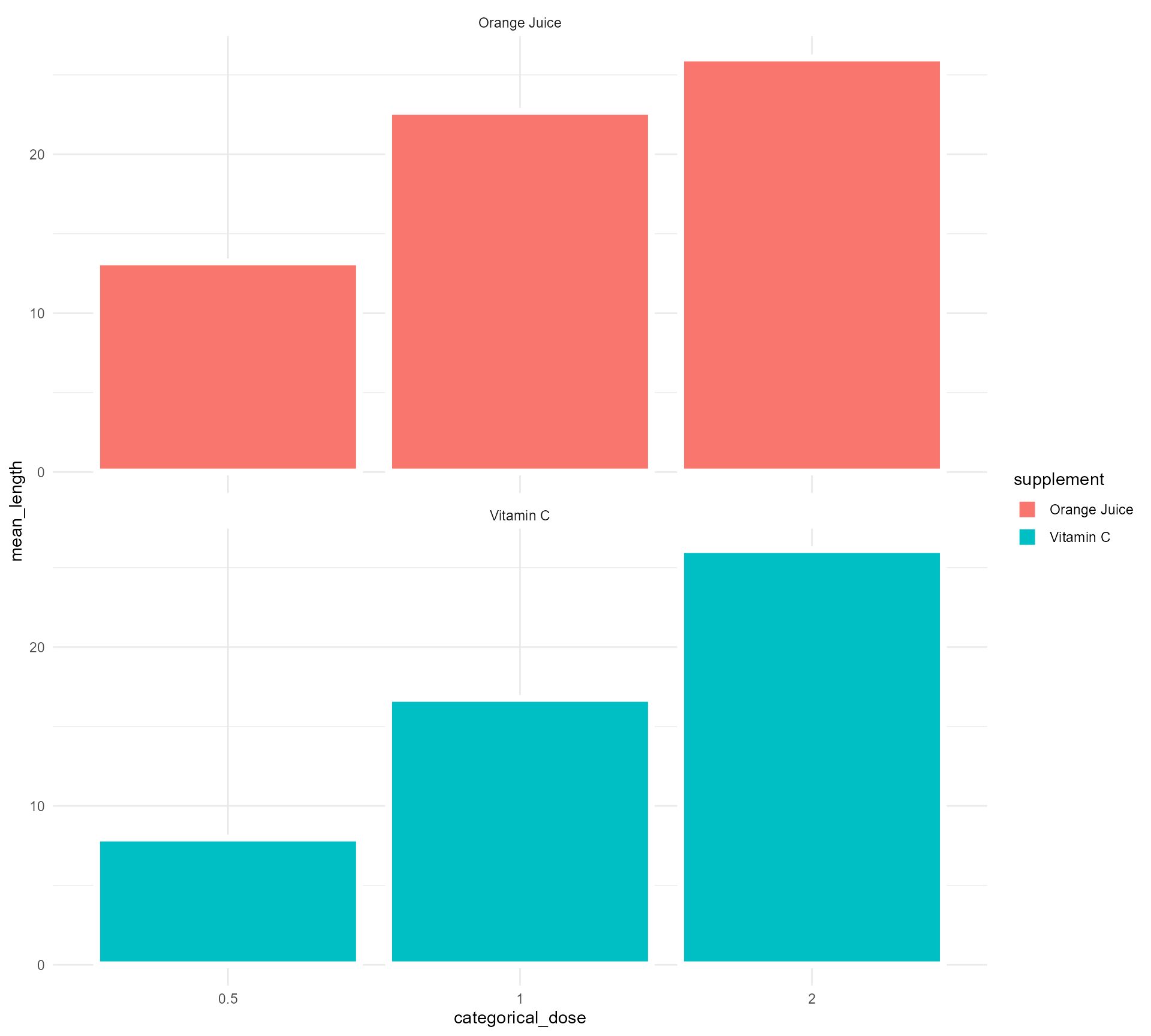
Turning Dose into a categorical variable (fear not!) + facetting
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
colour = "#FFFFFF",
size = 2) +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()Setting up our first plot
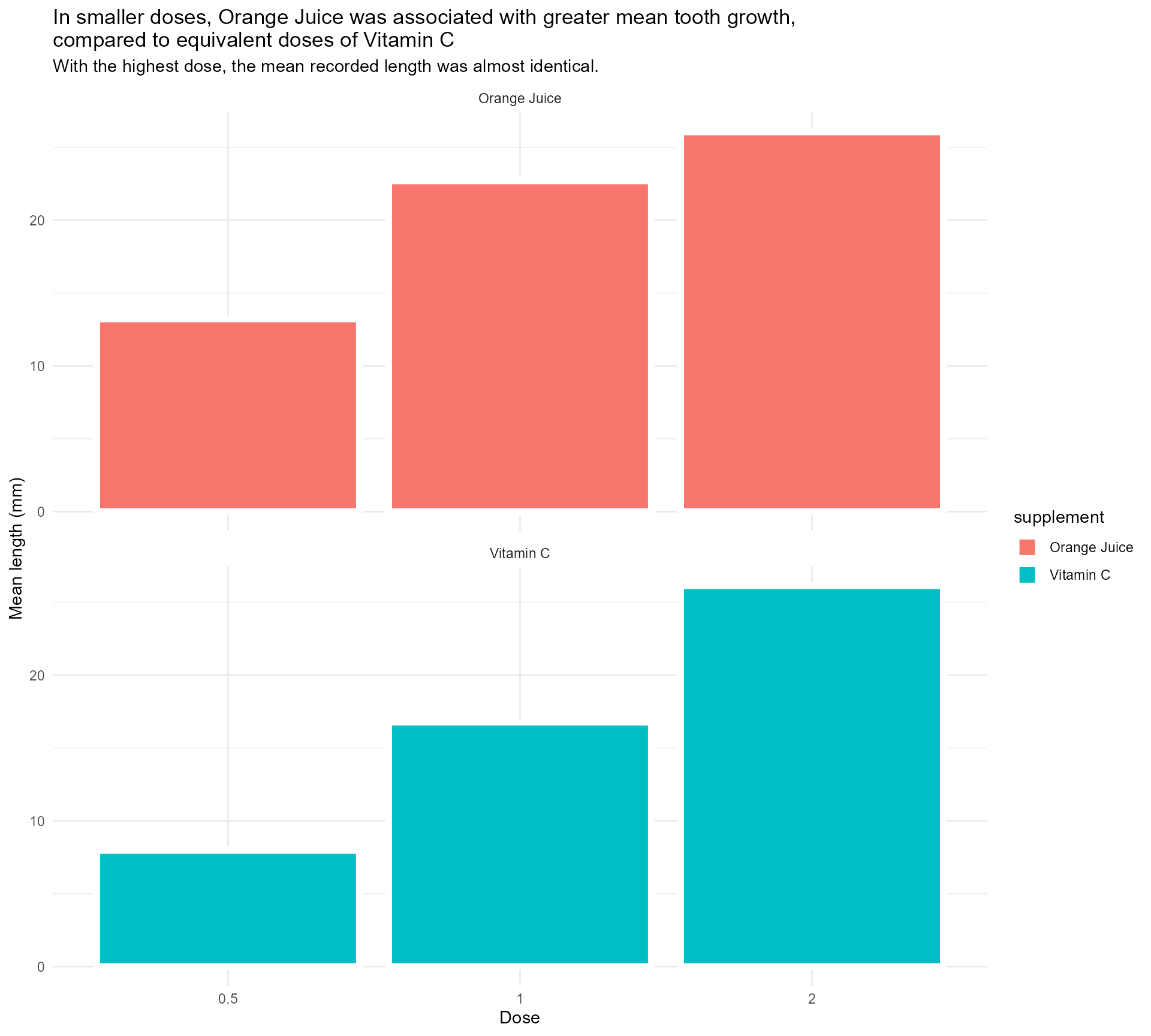
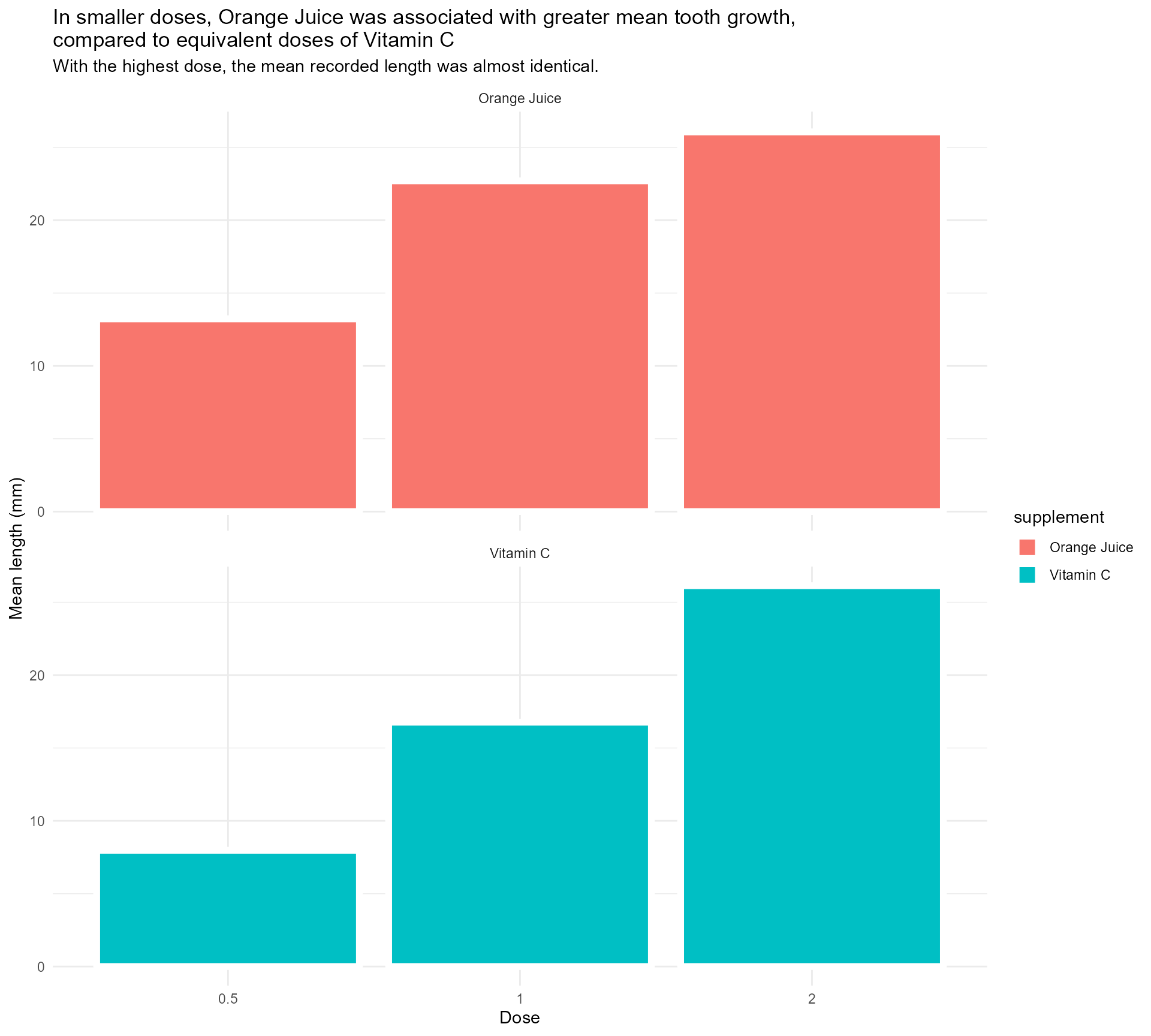
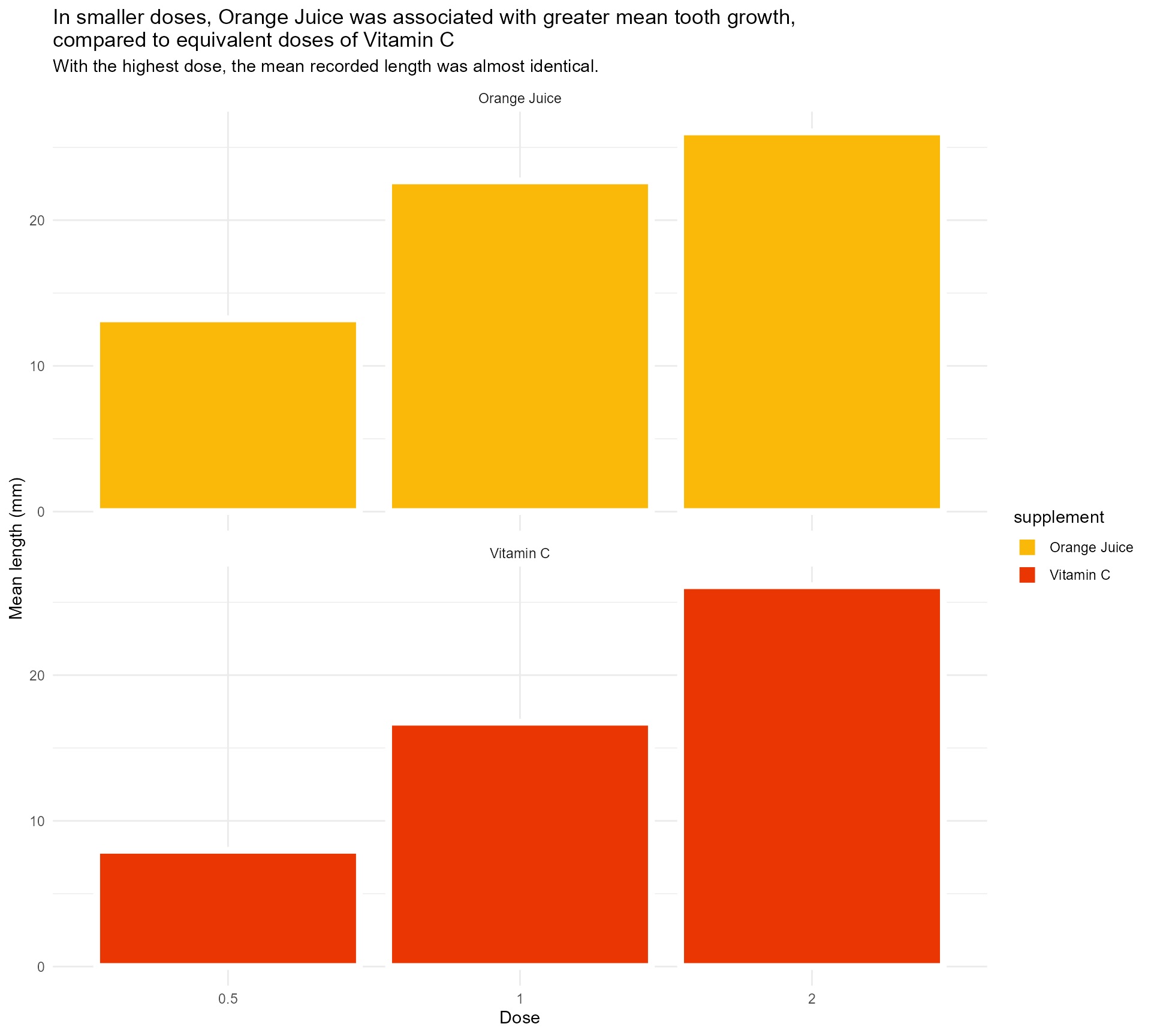
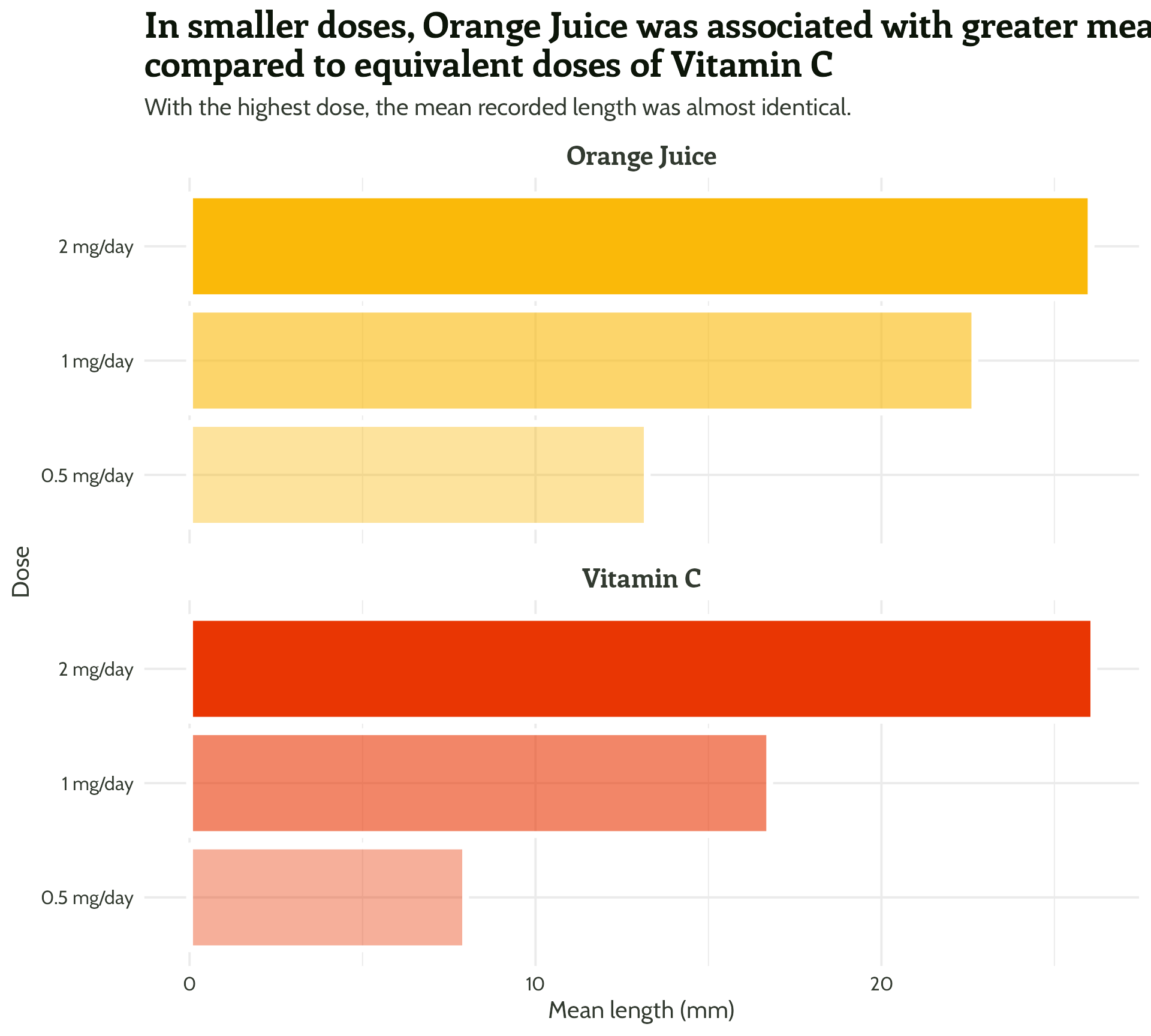
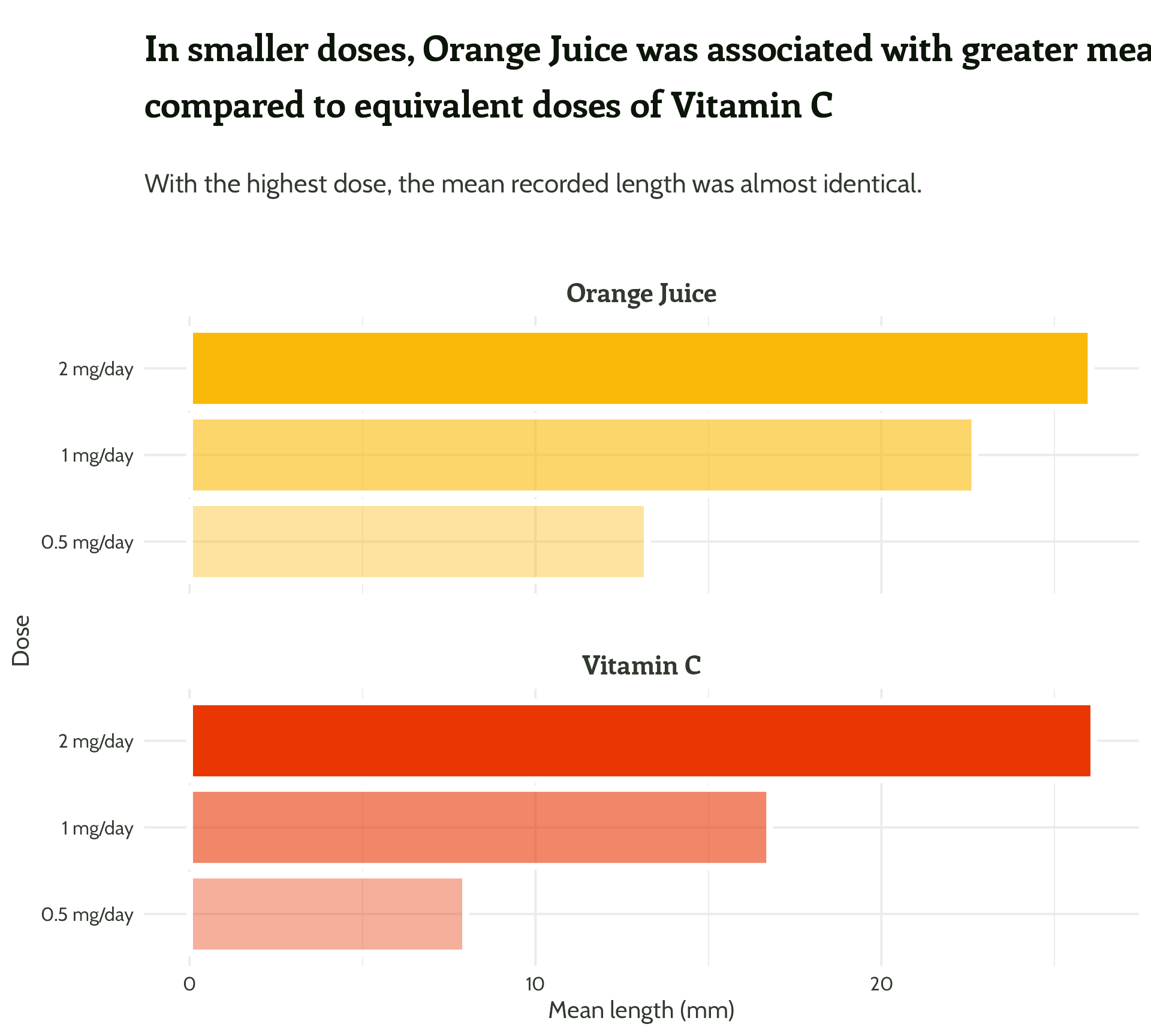
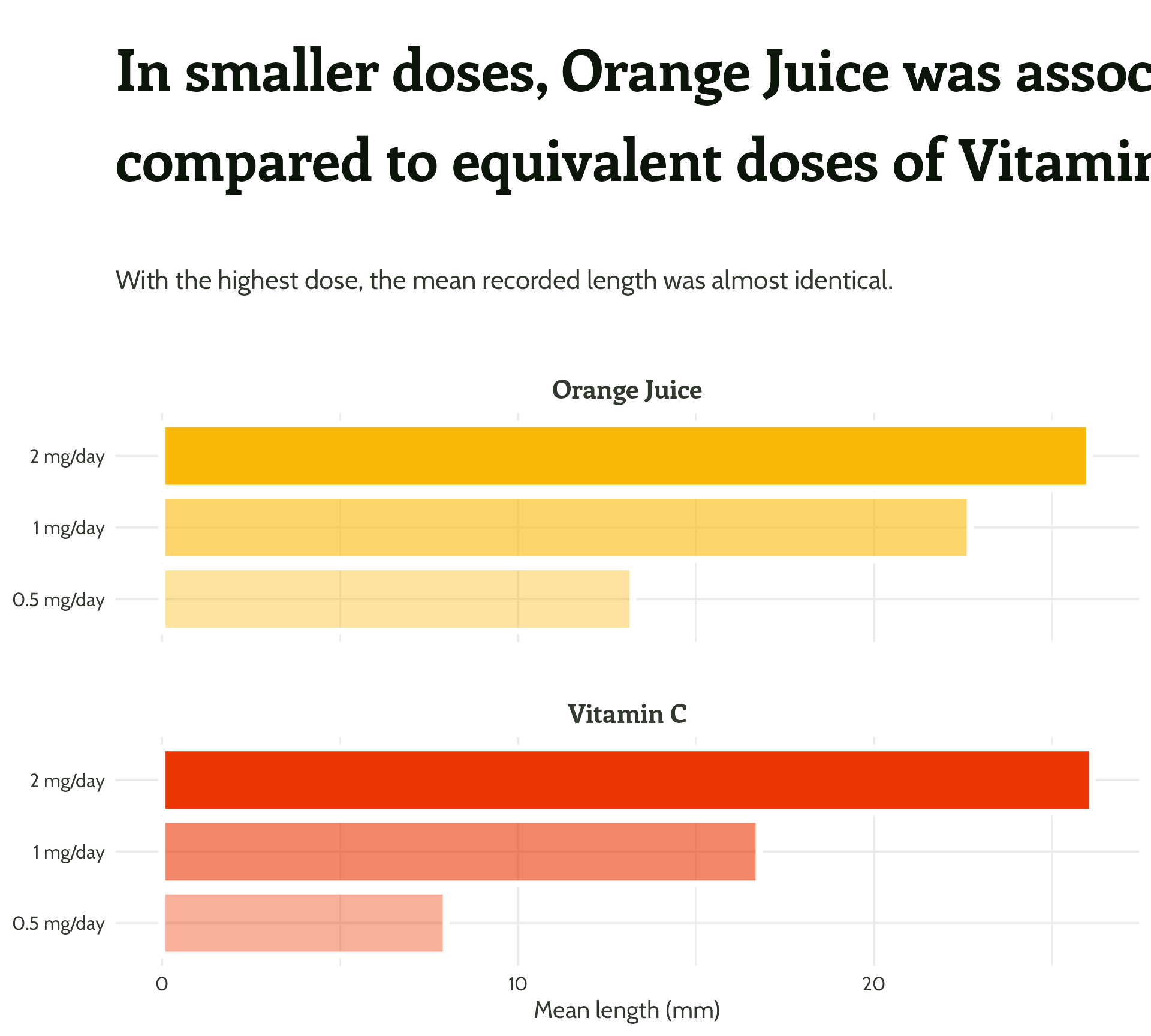
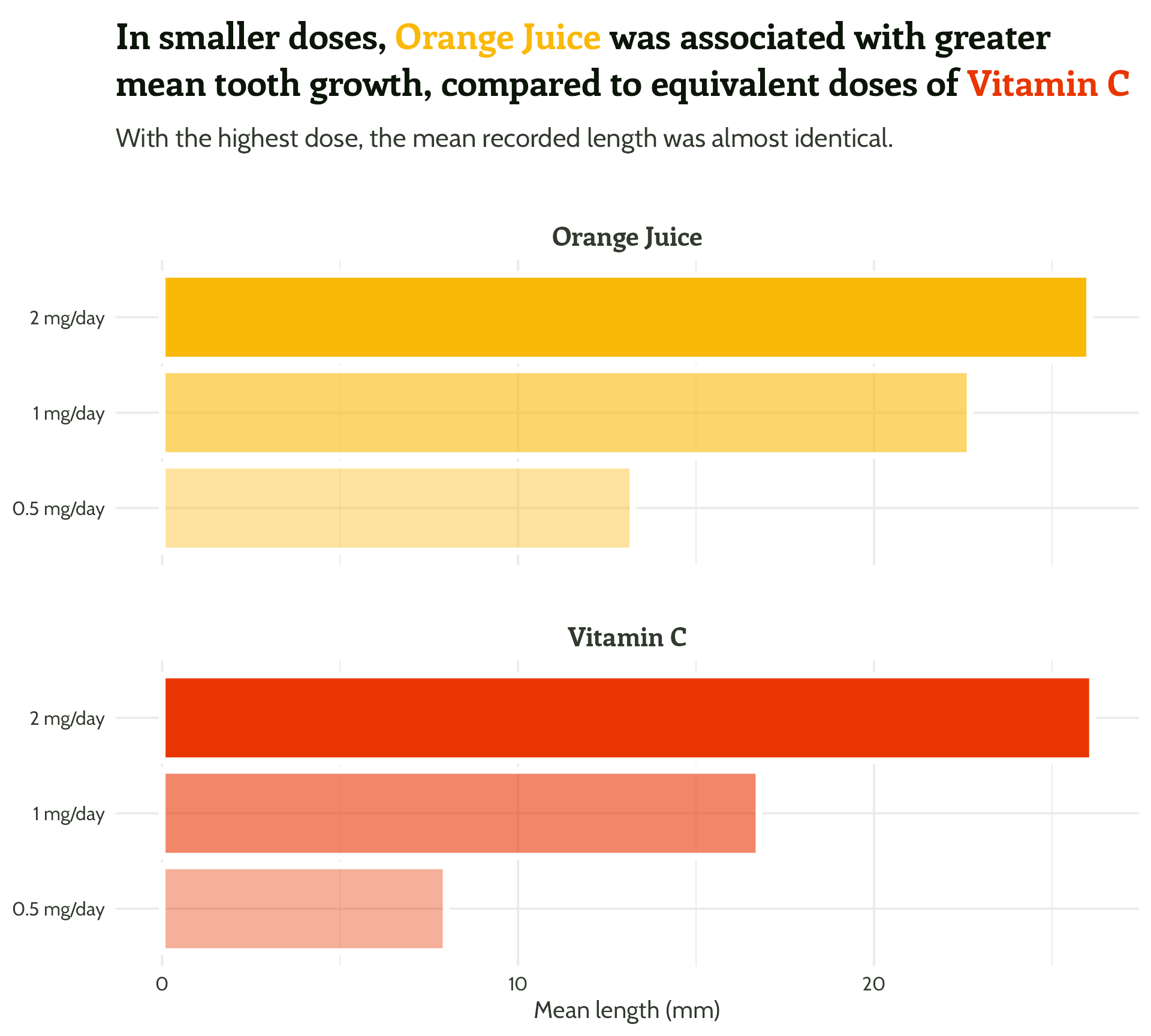
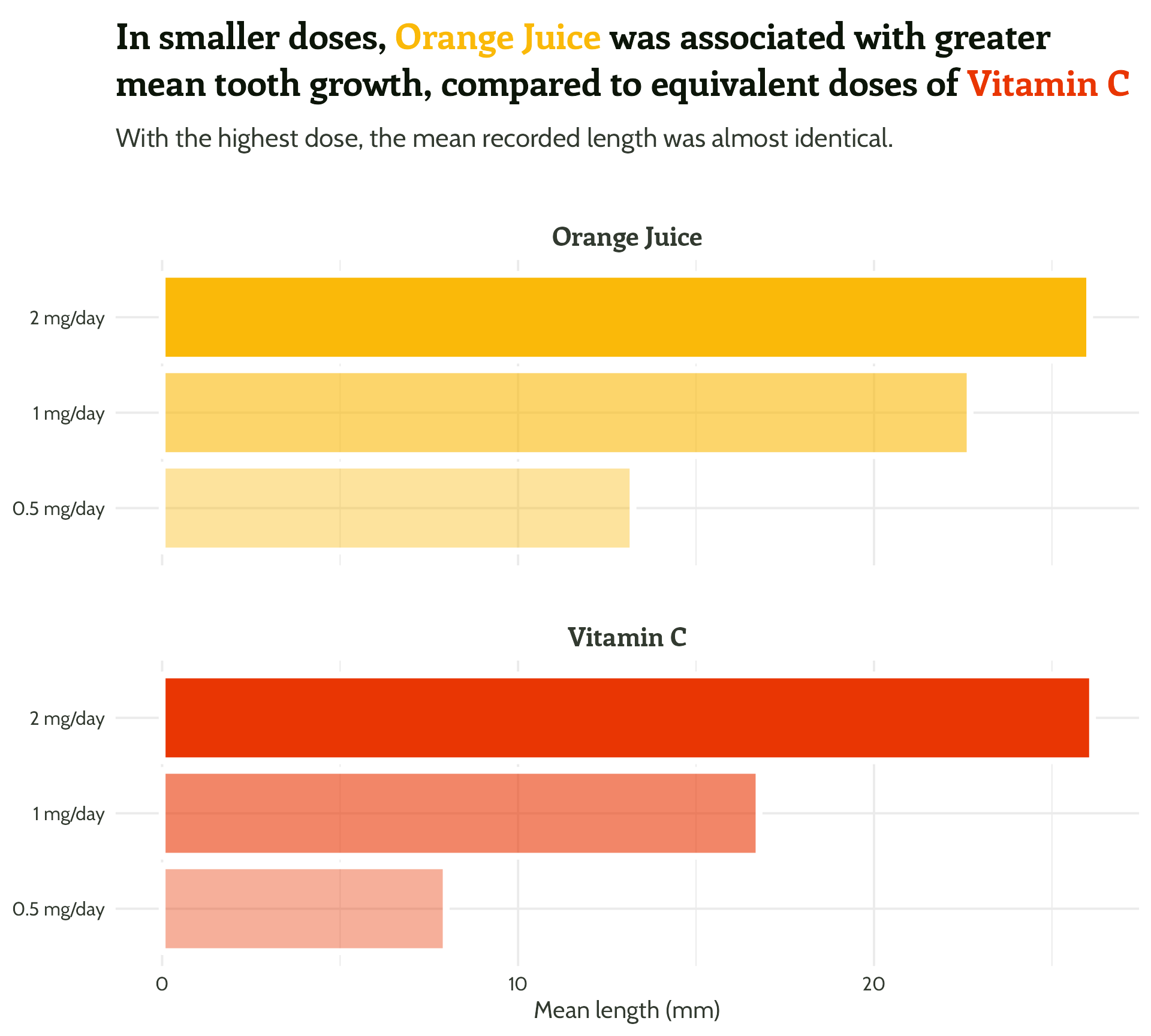
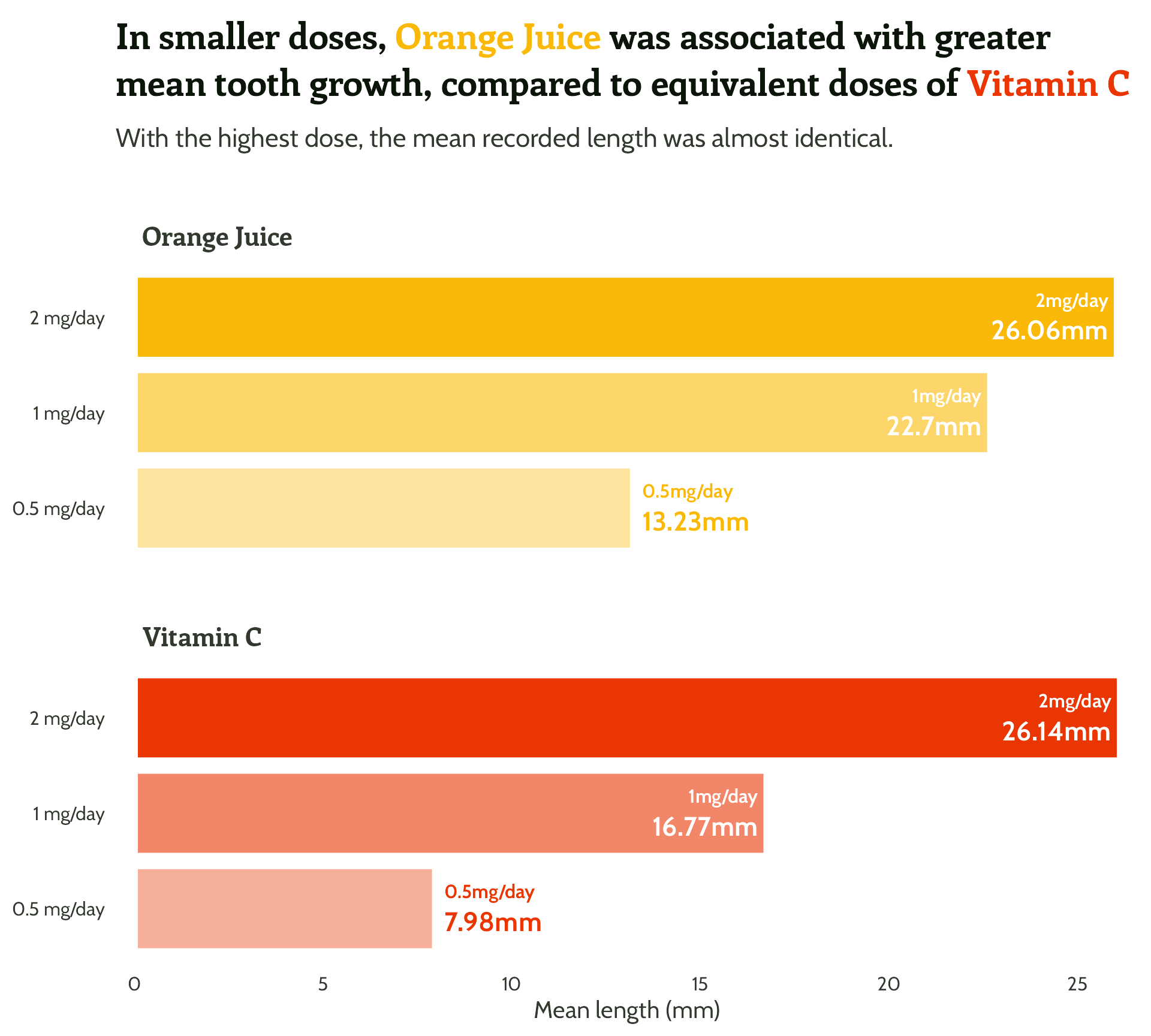
Adding some text (finally!)
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
colour = "#FFFFFF",
size = 2) +
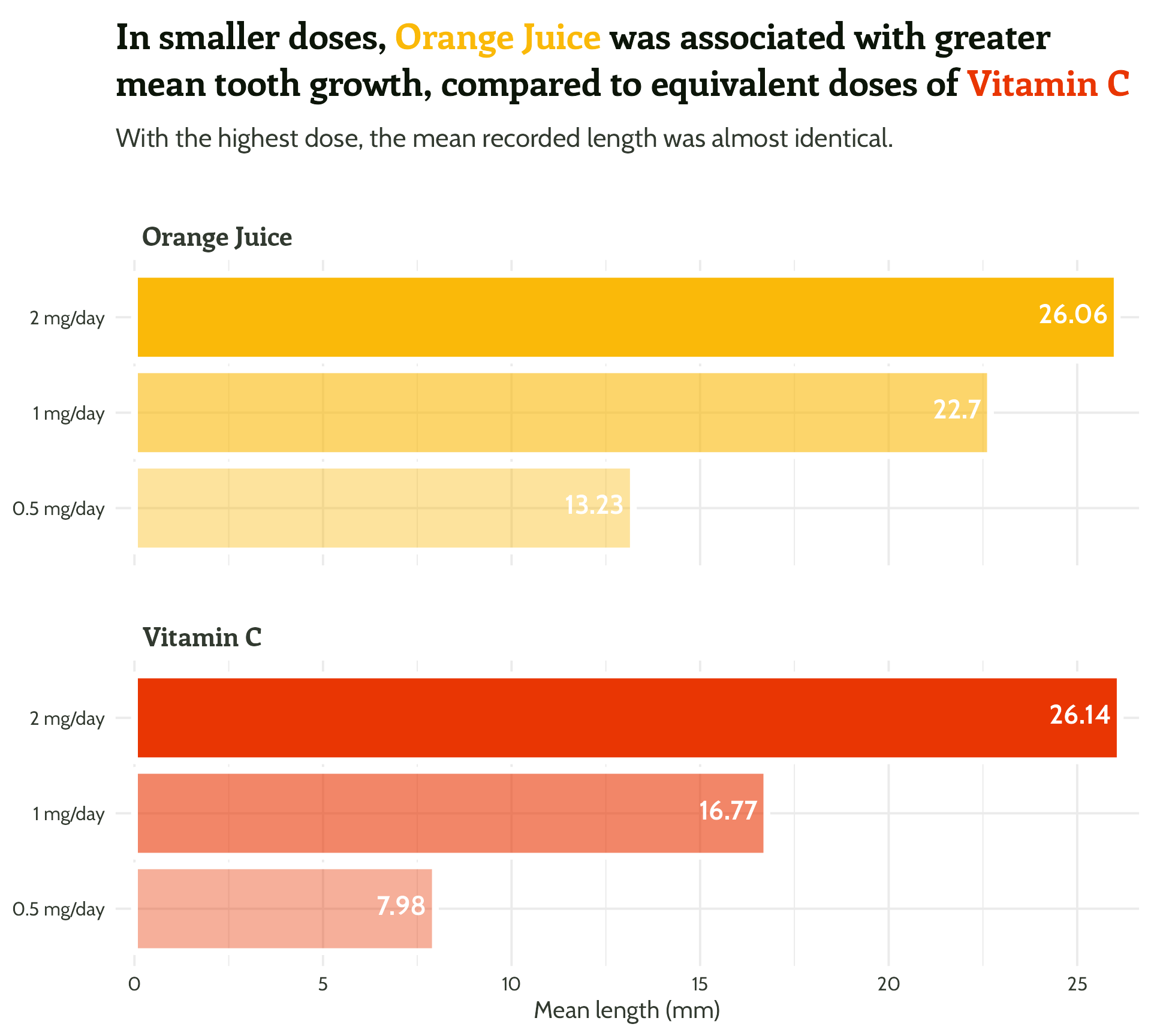
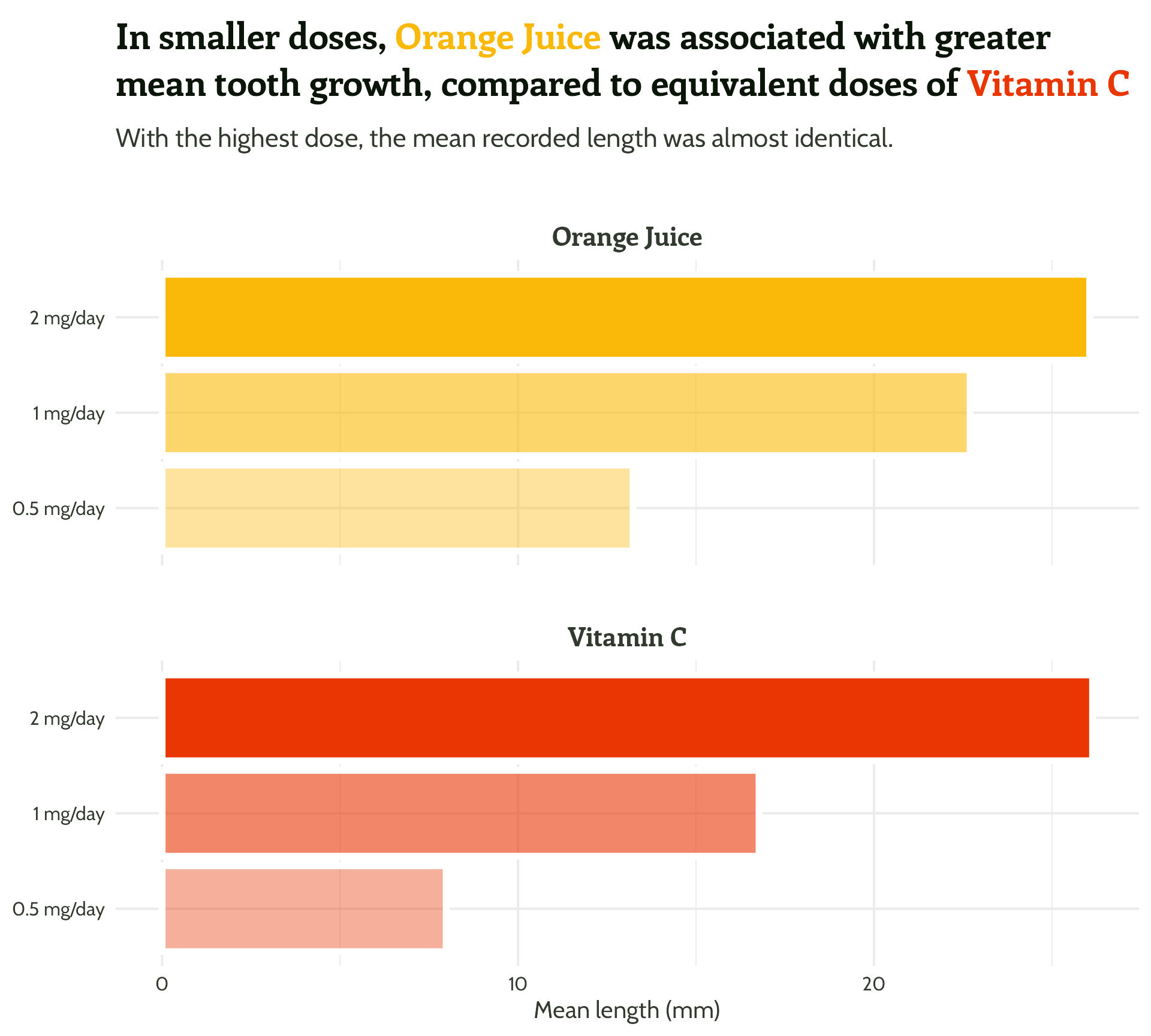
labs(x = "Dose",
y = "Mean length (mm)",
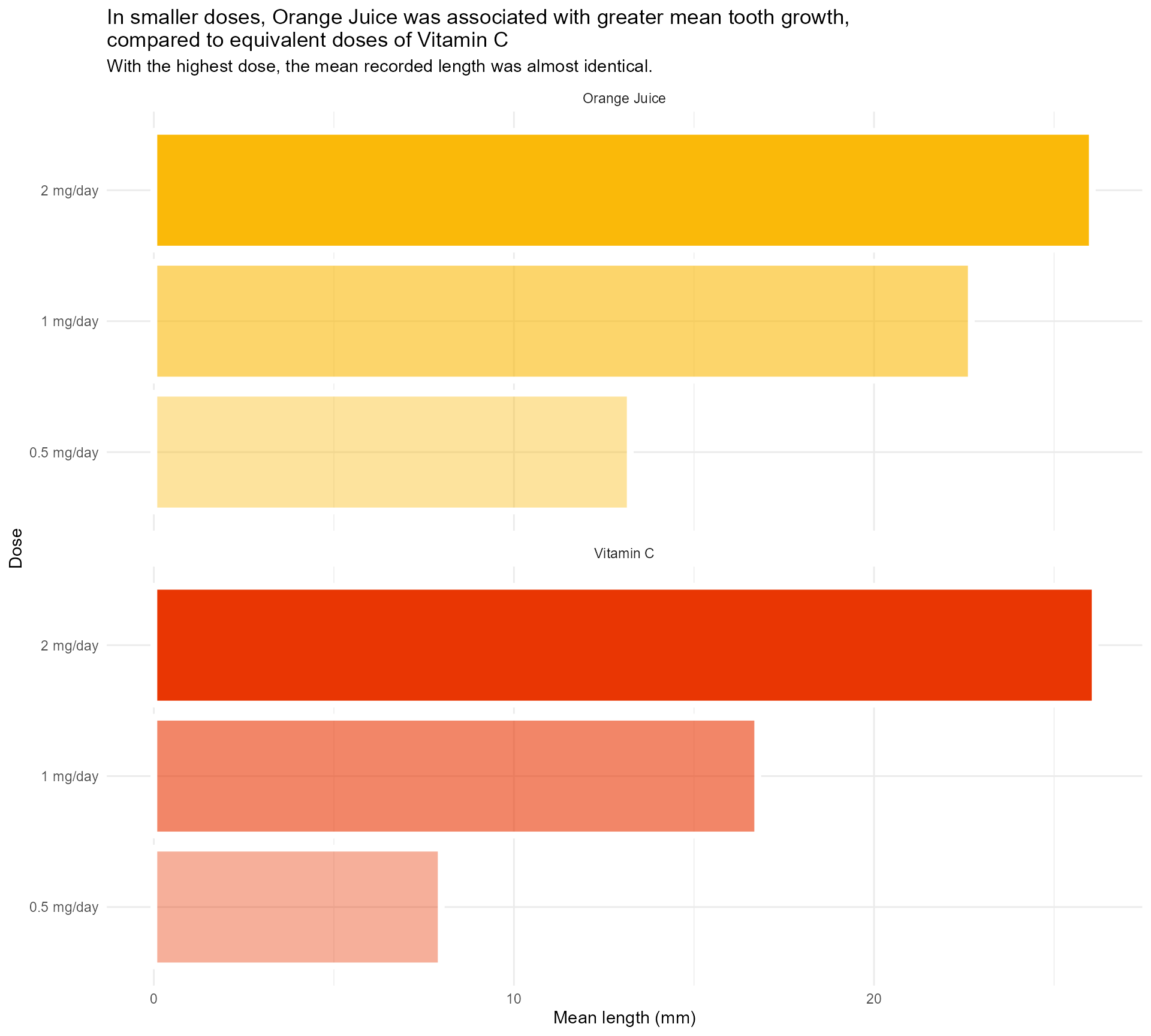
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()Setting up our first plot
Legend + facet strip + colour + title… Wait, which one is which?
#1 - Use colour purposefully
- Orange juice is… orange!
- Vitamin C is… also orange, but more red and “aggressive”
- Those green leaves look nice with those colours…
- imagecolorpicker.com
#1 - Use colour purposefully
Generating a colour palette, starting with orange juice! #fab909
[1] "#DB5A05" "#E93603" "#F71201"[1] "#3C6B30" "#0C1509"[1] "#0C1509" "#323A30" "#595F57" "#80857F" "#A7AAA6" "#CED0CD"#1 - Use colour purposefully
Creating a named vector

#1 - Use colour purposefully
Back to the plot!
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
colour = "#FFFFFF",
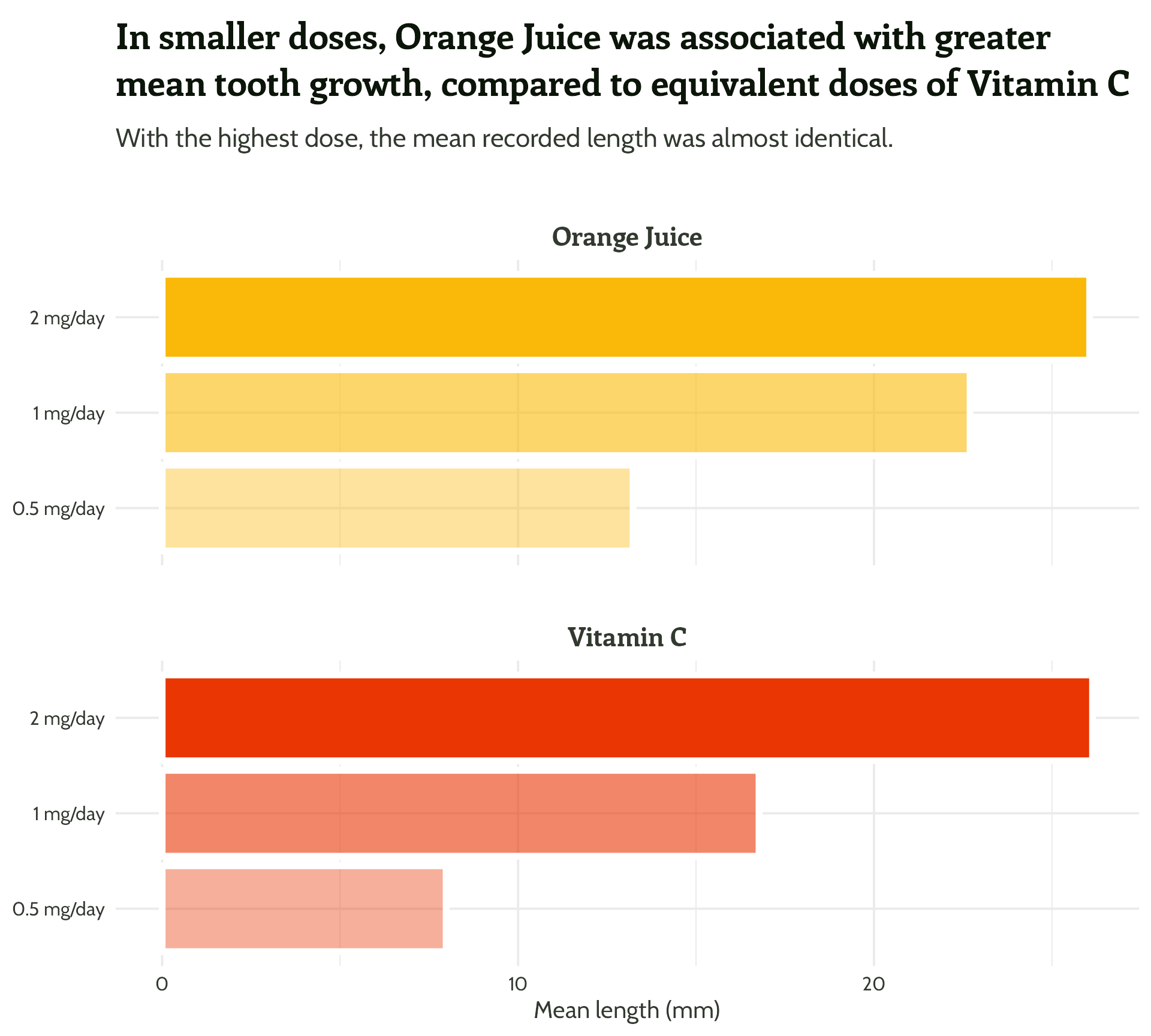
size = 2) +
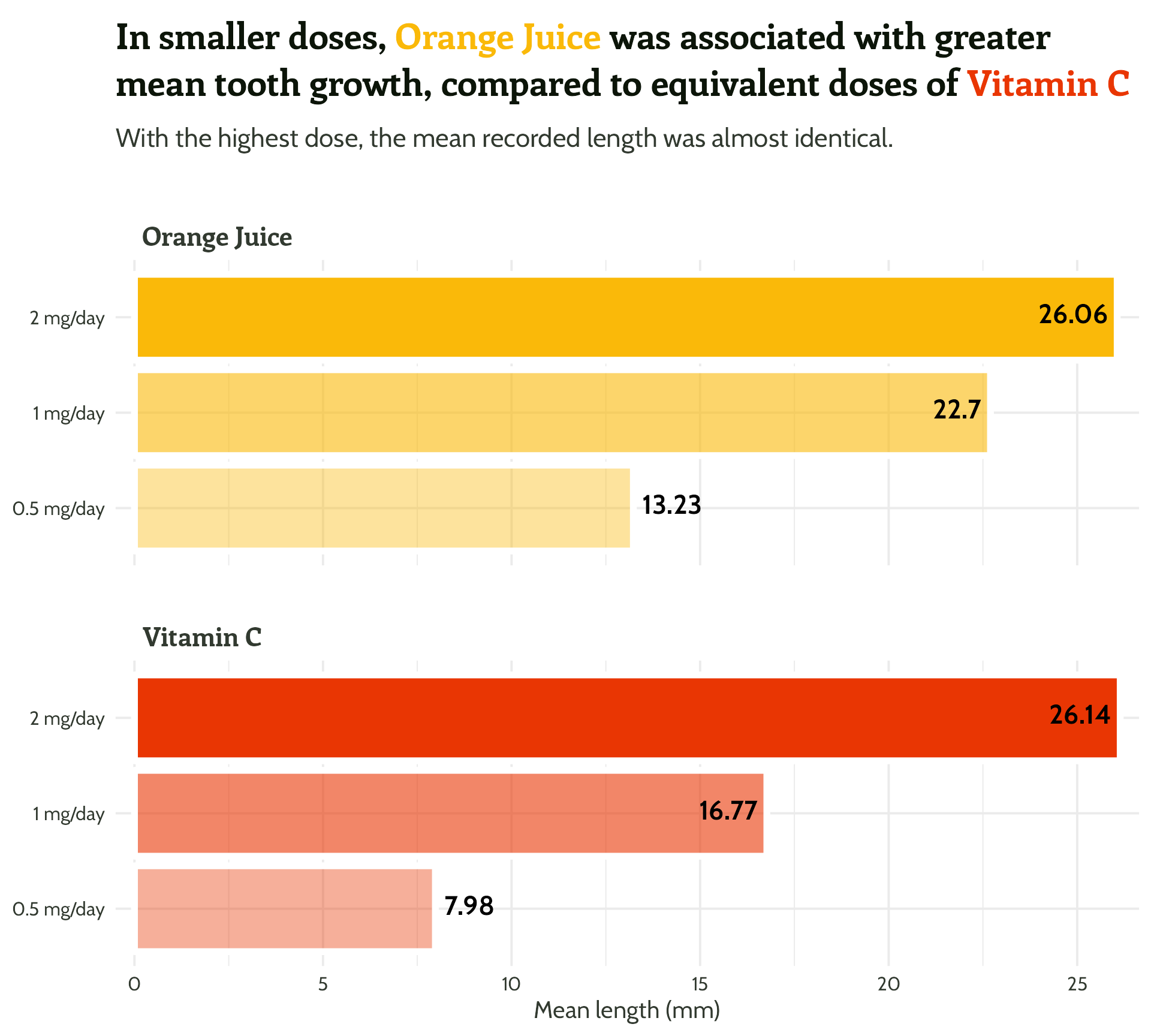
labs(x = "Dose",
y = "Mean length (mm)",
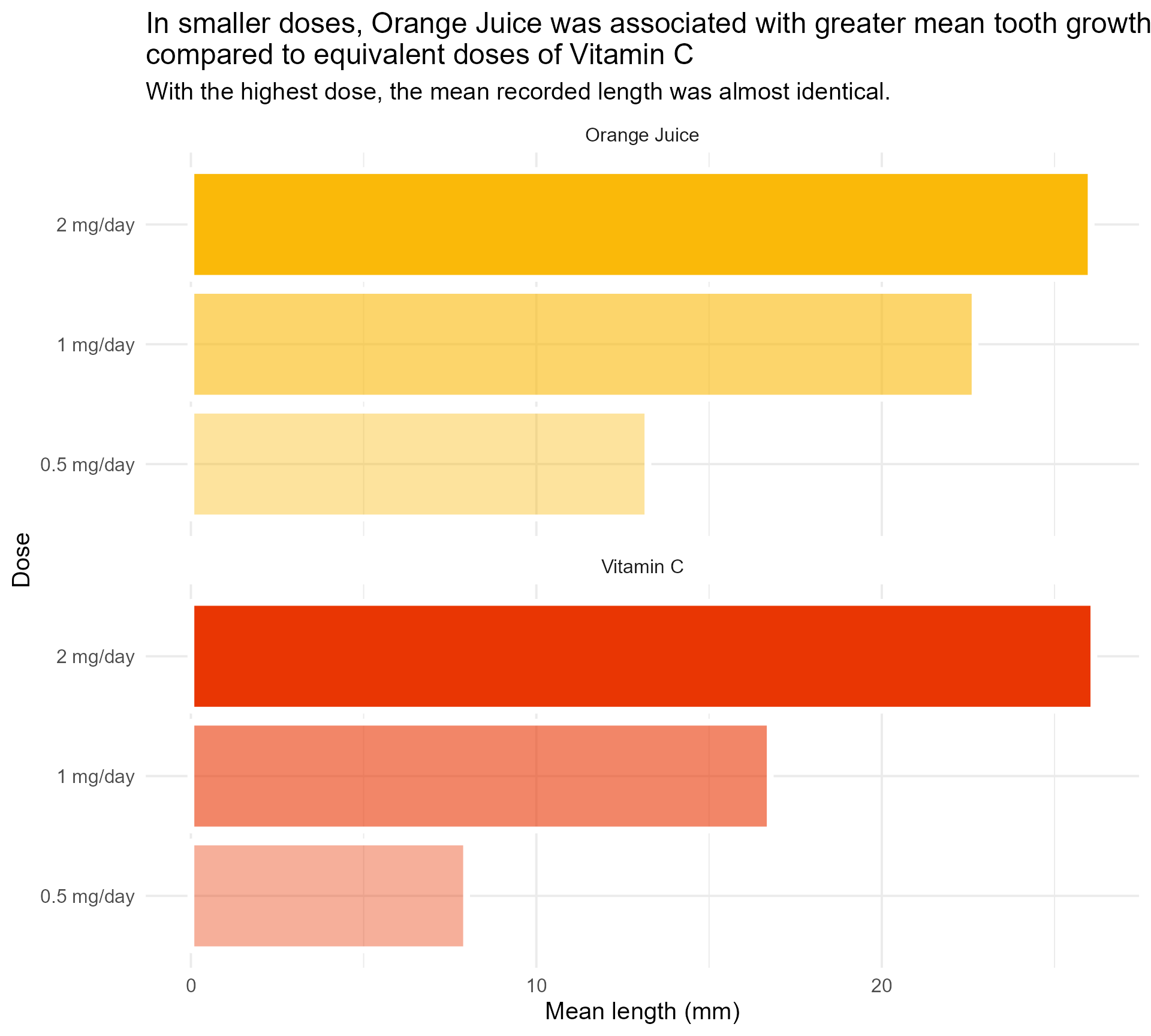
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()#1 - Use colour purposefully
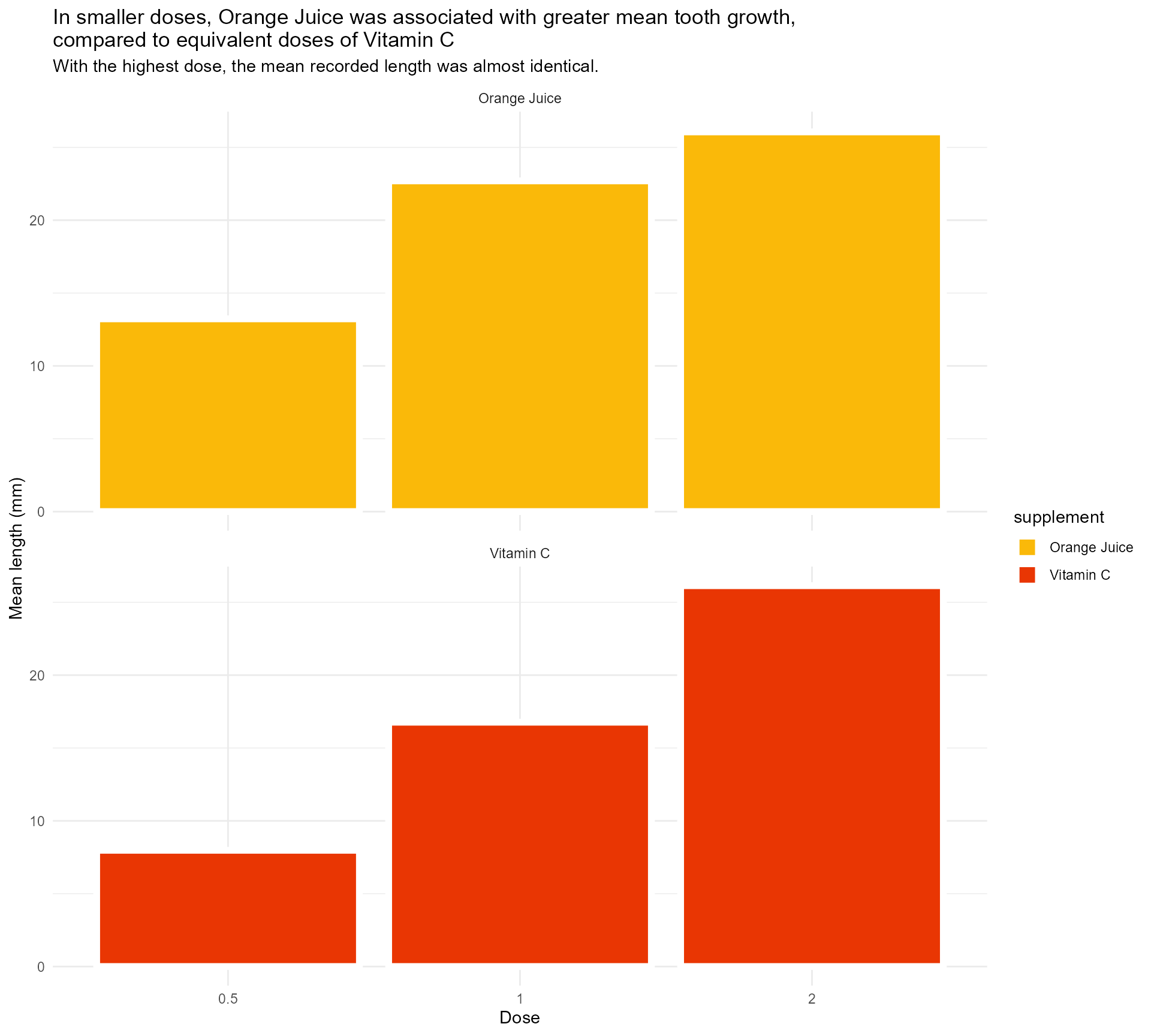
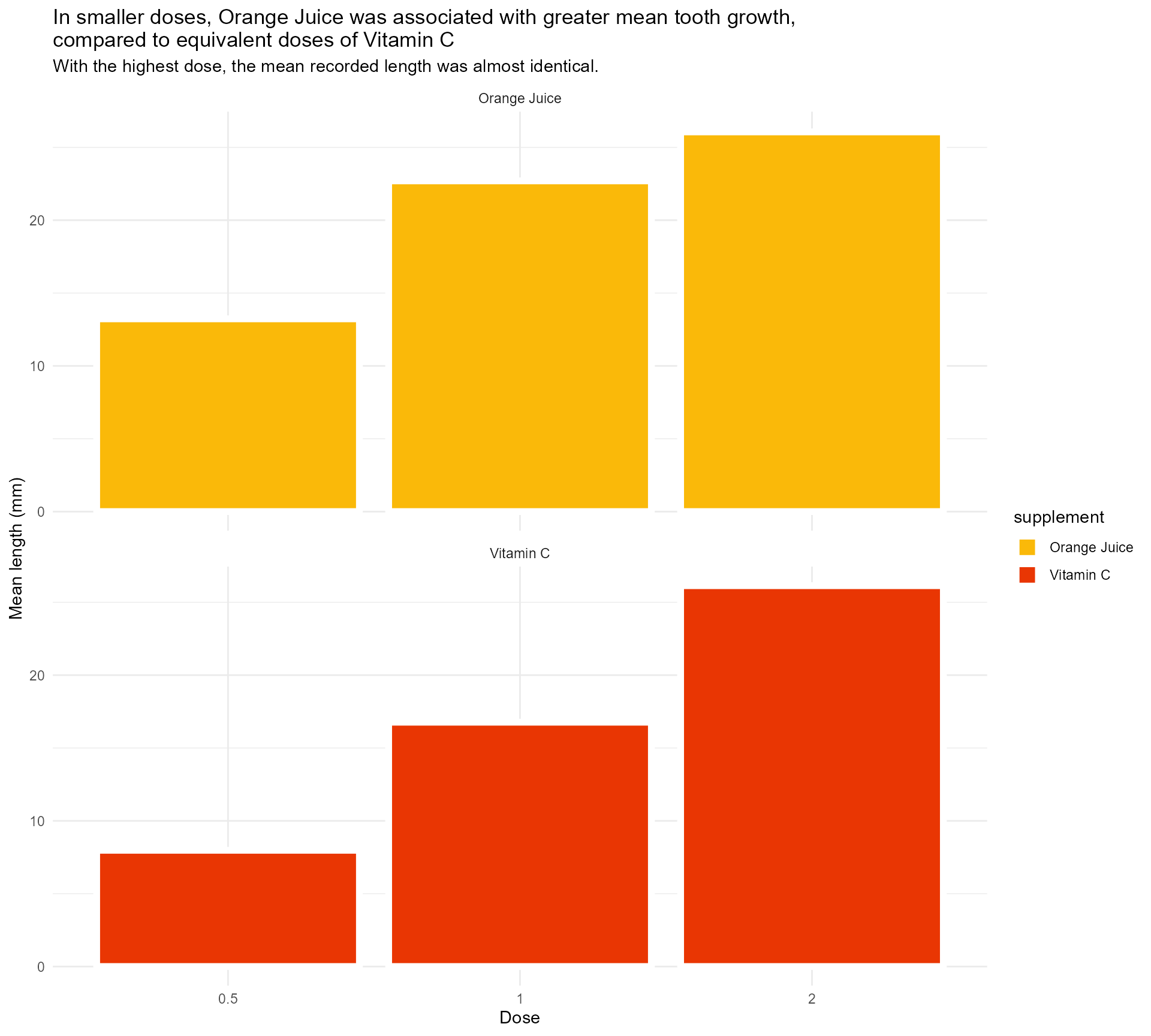
Add in our colours
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
colour = "#FFFFFF",
size = 2) +
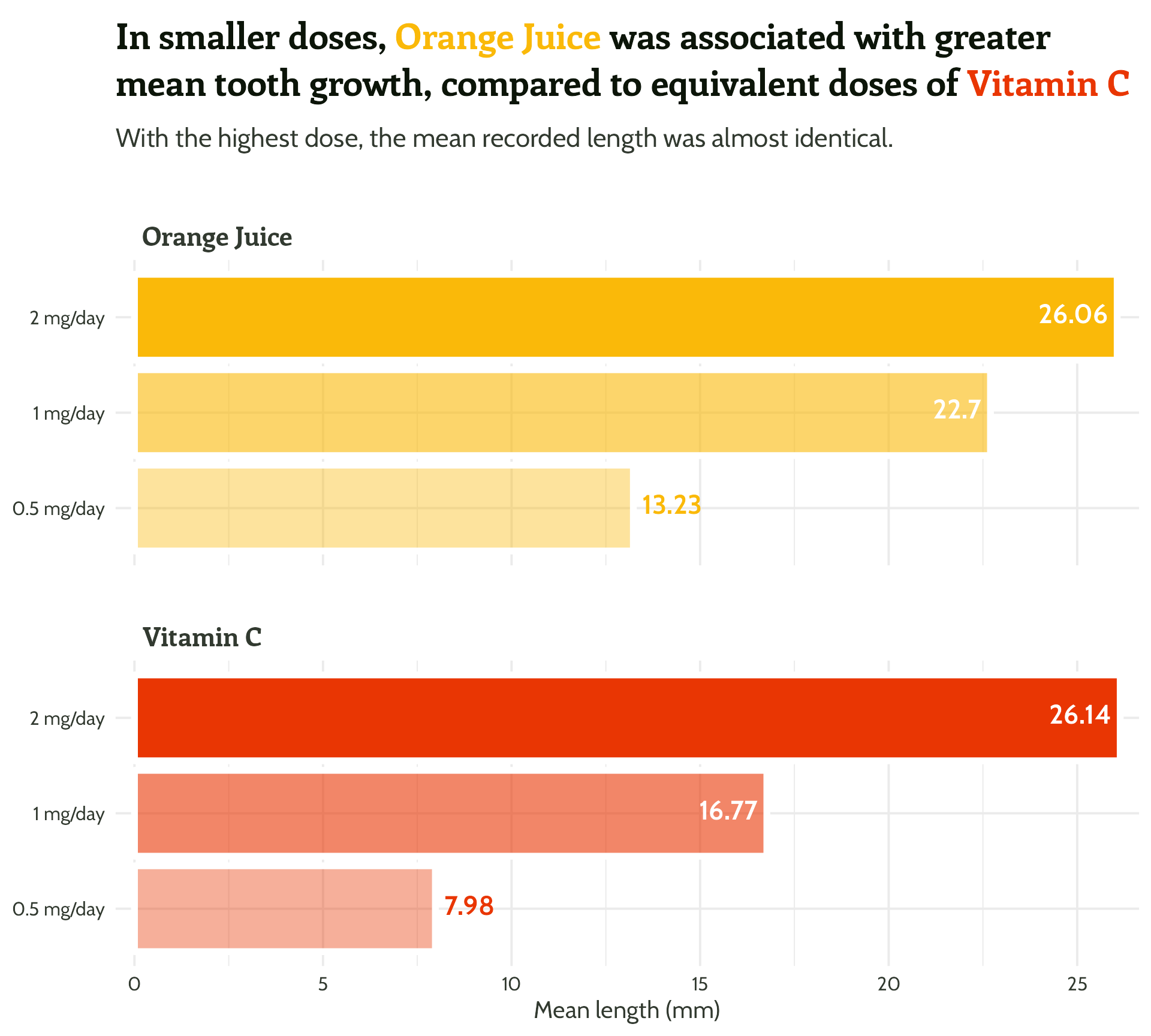
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = c("#fab909",
"#E93603")) +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()#1 - Use colour purposefully
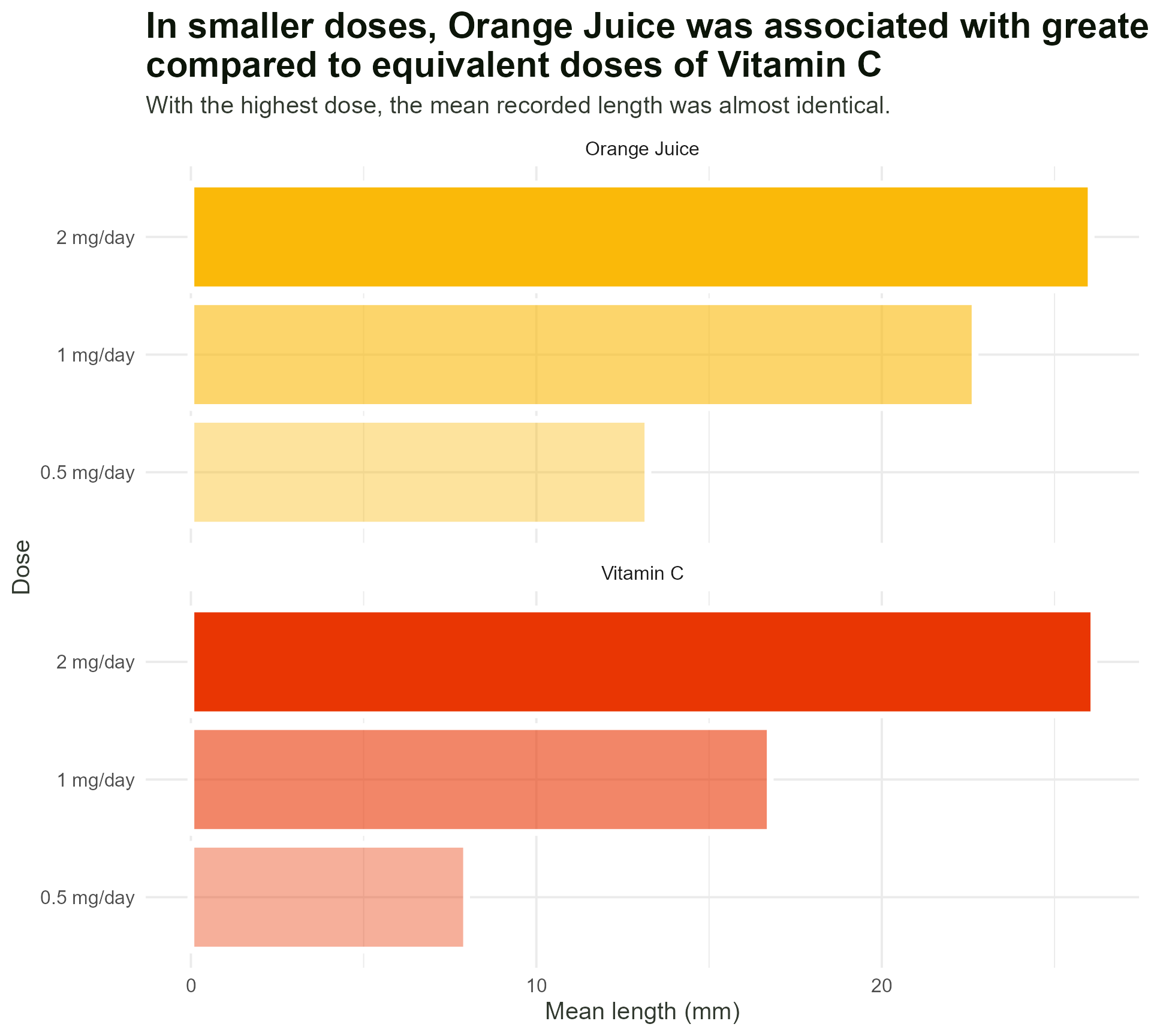
Add in our colours - wait, what?
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose),
supplement =
factor(supplement,
levels = c("Vitamin C",
"Orange Juice"))) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
colour = "#FFFFFF",
size = 2) +
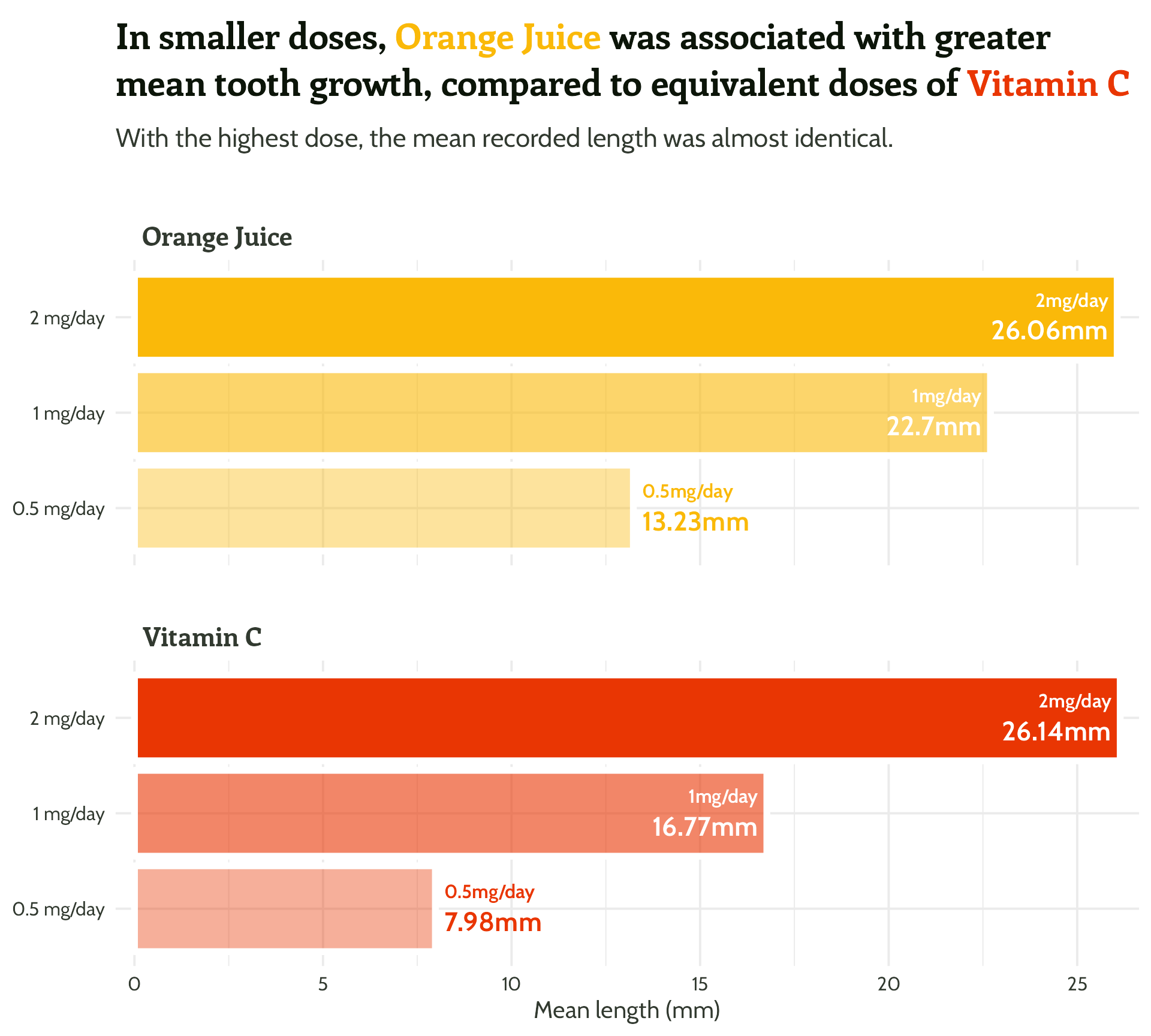
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = c("#fab909",
"#E93603")) +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()#1 - Use colour purposefully
Add in our colours - named vector to the rescue!
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose),
supplement =
factor(supplement,
levels = c("Vitamin C",
"Orange Juice"))) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
colour = "#FFFFFF",
size = 2) +
labs(x = "Dose",
y = "Mean length (mm)",
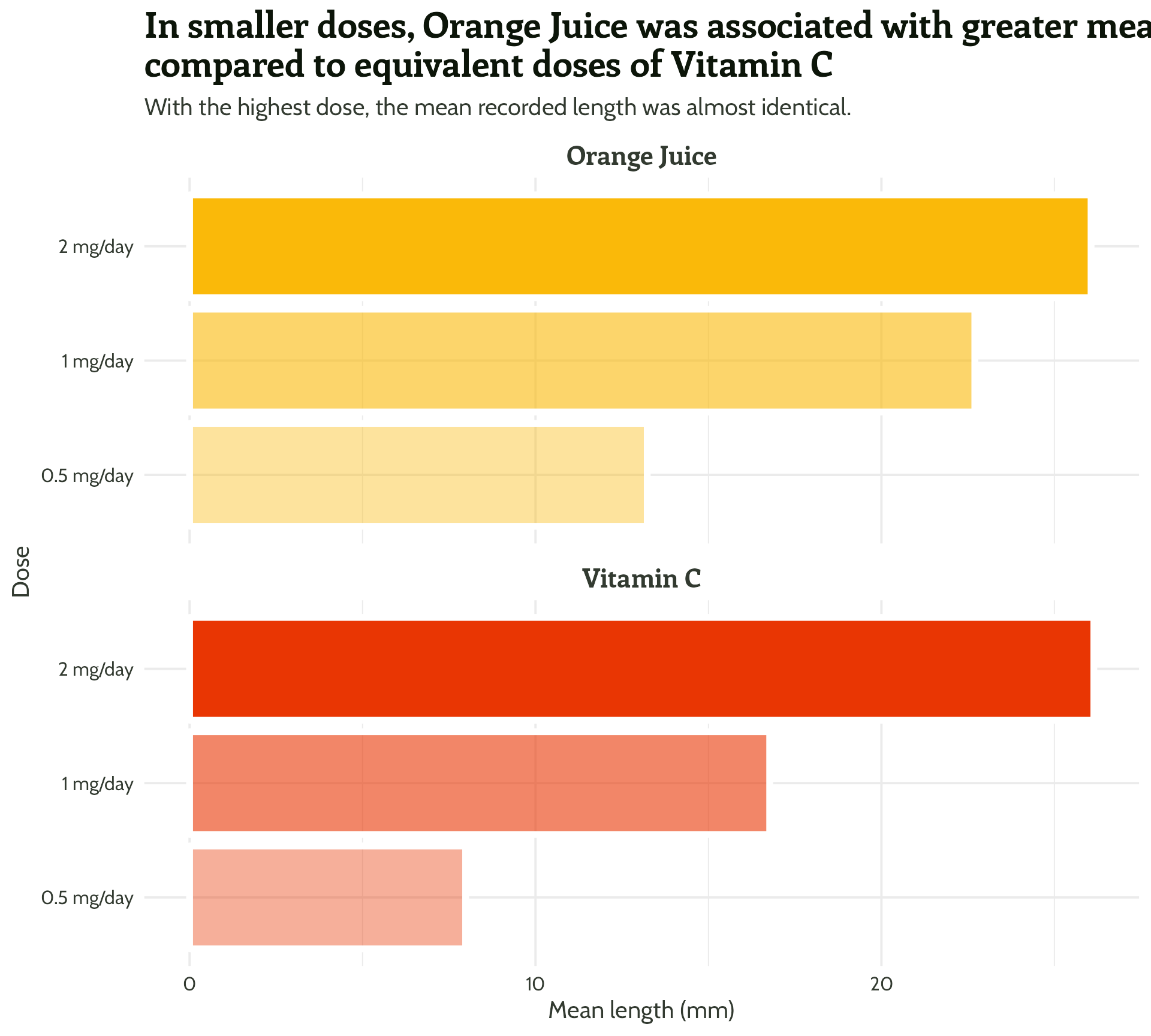
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = vit_c_palette) +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()#1 - Use colour purposefully
Add in our colours - named vector to the rescue!
Key advantages
- Know the colours are applied to the right data points!
- Keep colour-data pairings consistent throughout the project
- Package up a default palette
- Easily reuse colours in the text
ggtext::element_markdown()later in this workshop
#1 - Use colour purposefully
Get rid of unused colours
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
colour = "#FFFFFF",
size = 2) +
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = vit_c_palette) +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()#1 - Use colour purposefully
Get rid of unused colours
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(stat = "identity",
colour = "#FFFFFF",
size = 2) +
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = vit_c_palette,
limits = force) +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()#1 - Use colour purposefully
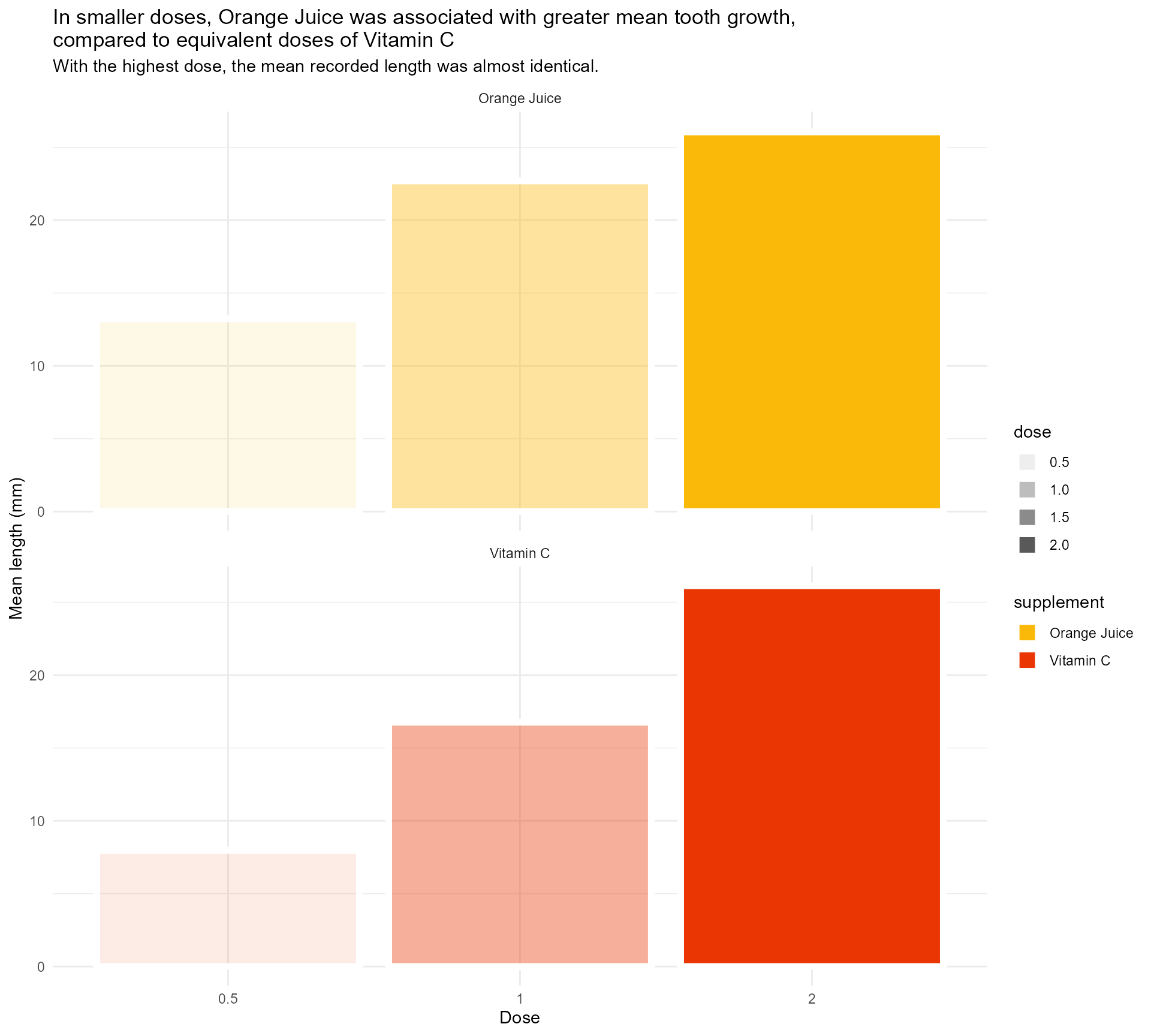
Use transparency to indicate dose
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(aes(alpha = dose),
stat = "identity",
colour = "#FFFFFF",
size = 2) +
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = vit_c_palette, limits = force) +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()#1 - Use colour purposefully
Use transparency to indicate dose - within limits
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(aes(alpha = dose),
stat = "identity",
colour = "#FFFFFF",
size = 2) +
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = vit_c_palette, limits = force) +
scale_alpha(range = c(0.33, 1)) +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()#1 - Use colour purposefully
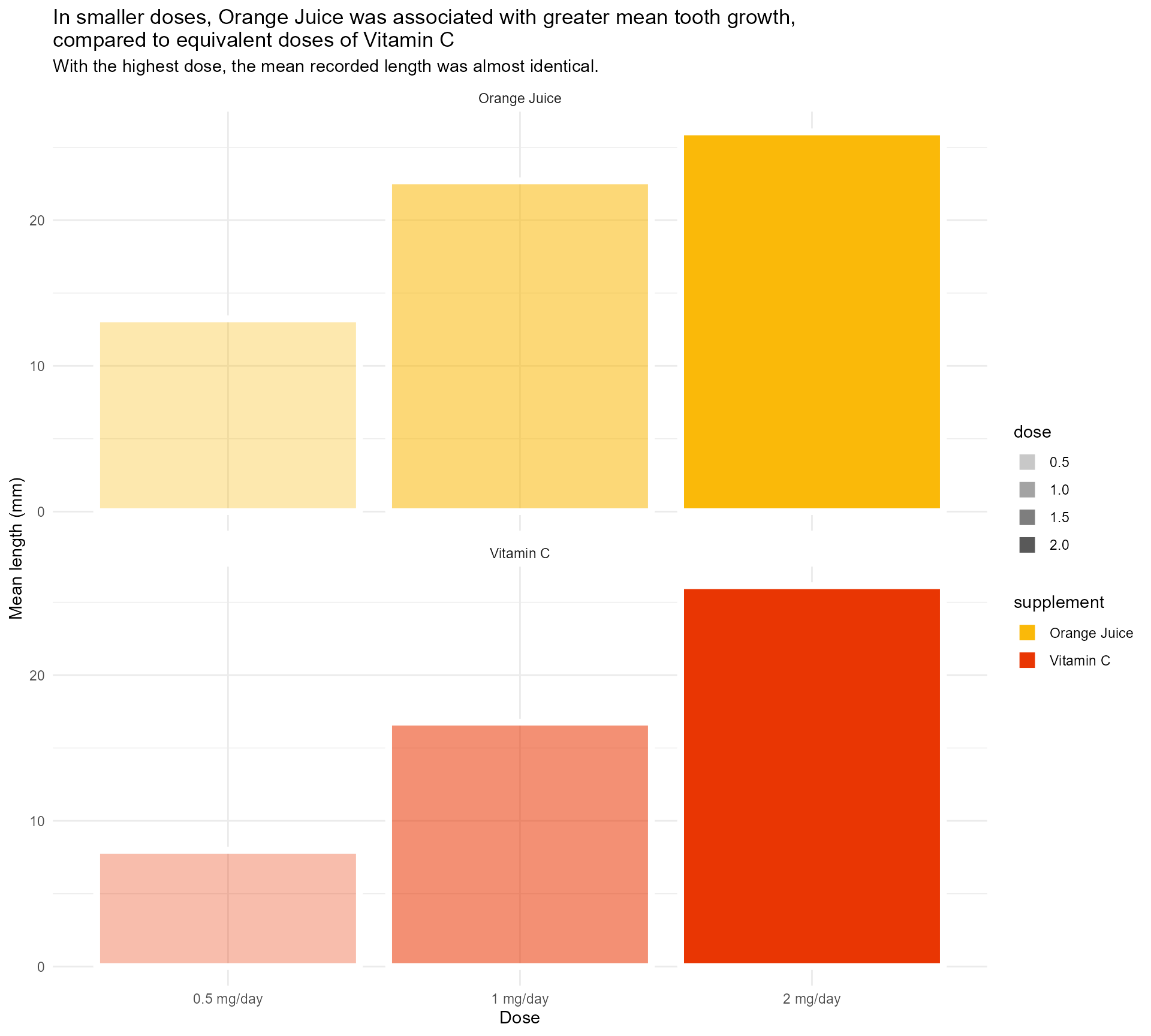
What is the dose unit again? ?ToothGrowth
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(aes(alpha = dose),
stat = "identity",
colour = "#FFFFFF",
size = 2) +
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = vit_c_palette, limits = force) +
scale_alpha(range = c(0.33, 1)) +
scale_x_discrete(breaks = c("0.5", "1", "2"),
labels = function(x)
paste0(x, " mg/day")) +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal()#1 - Use colour purposefully
Legend has always been redundant!
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(aes(alpha = dose),
stat = "identity",
colour = "#FFFFFF",
size = 2) +
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = vit_c_palette, limits = force) +
scale_alpha(range = c(0.33, 1)) +
facet_wrap(supplement ~ ., ncol = 1) +
scale_x_discrete(breaks = c("0.5", "1", "2"), labels = function(x) paste0(x, " mg/day")) +
theme_minimal() +
theme(legend.position = "none")#1 - Use colour (and orientation) purposefully
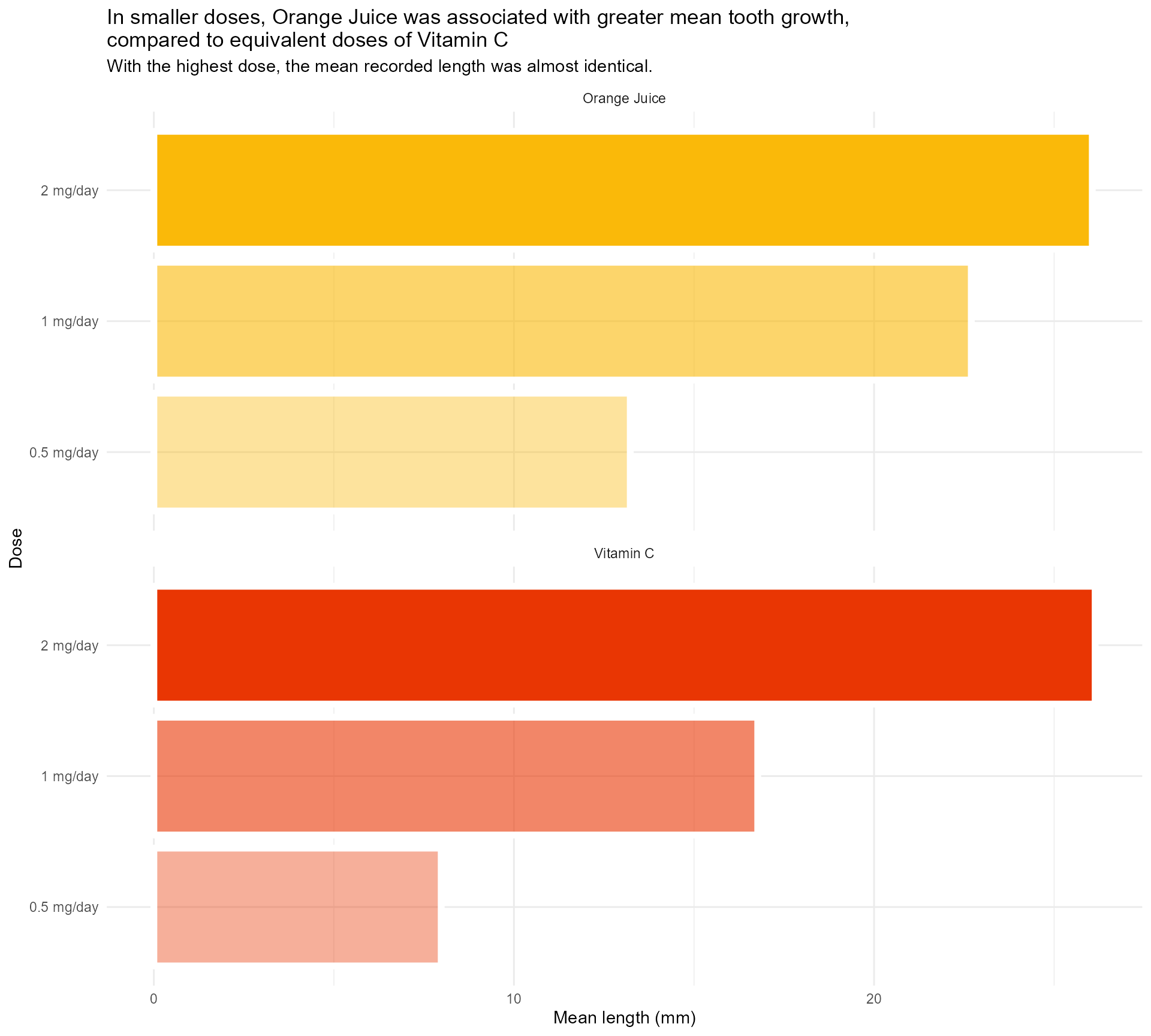
And I find this so much less confusing!
ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose,
y = mean_length,
fill = supplement)) +
geom_bar(aes(alpha = dose),
stat = "identity",
colour = "#FFFFFF",
size = 2) +
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = vit_c_palette, limits = force) +
scale_alpha(range = c(0.4, 1)) +
scale_x_discrete(breaks = c("0.5", "1", "2"), labels = function(x) paste0(x, " mg/day")) +
coord_flip() +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal() +
theme(legend.position = "none")#1 - Use colour (and orientation) purposefully
So much clearer, and we haven’t even done any annotating!
Five tips for beautiful annotations
#1 - Use colour purposefully
#2 - Add text hierarchy
#3 - Reduce unnecessary eye movement
#4 - Highlight important patterns
#5 - See how much you can declutter
Bonus track - {gghighlight}, Tee pipe and curved arrows
#2 - Add text hierarchy
Find out more: https://www.interaction-design.org/
#2 - Add text hierarchy
Time to start playing with theme()!
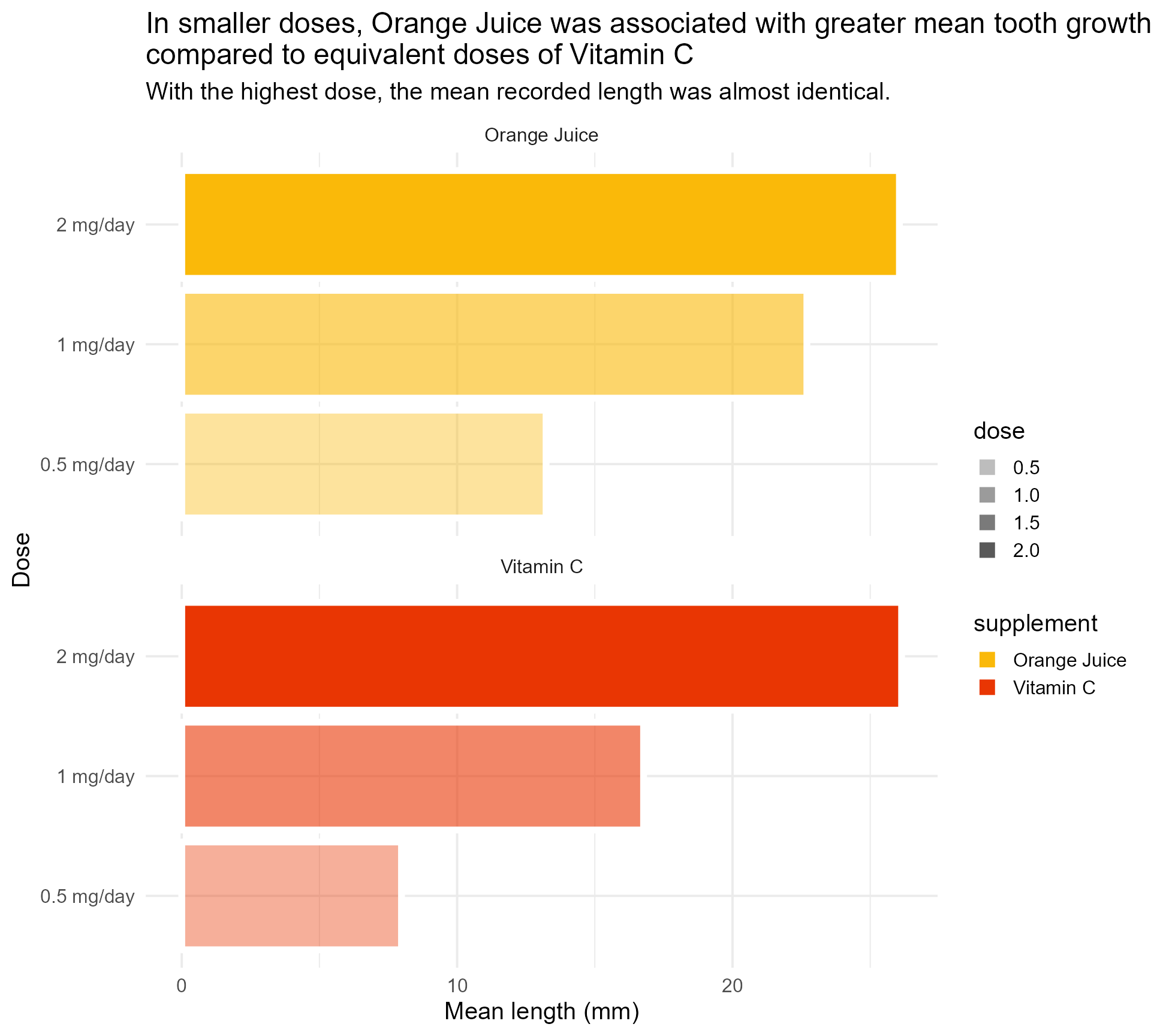
basic_plot <- ToothGrowth %>%
mutate(supplement = case_when(supp == "OJ" ~ "Orange Juice", supp == "VC" ~ "Vitamin C", TRUE ~ as.character(supp))) %>%
group_by(supplement, dose) %>%
summarise(mean_length = mean(len)) %>%
mutate(categorical_dose = factor(dose)) %>%
ggplot(aes(x = categorical_dose, y = mean_length, fill = supplement)) +
geom_bar(aes(alpha = dose), stat = "identity", colour = "#FFFFFF", size = 2) +
labs(x = "Dose",
y = "Mean length (mm)",
title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C",
subtitle = "With the highest dose, the mean recorded length was almost identical.") +
scale_fill_manual(values = vit_c_palette, limits = force) +
scale_alpha(range = c(0.4, 1)) +
scale_x_discrete(breaks = c("0.5", "1", "2"), labels = function(x) paste0(x, " mg/day")) +
coord_flip() +
facet_wrap(supplement ~ ., ncol = 1) +
theme_minimal(base_size = 15)
basic_plot#2 - Add text hierarchy
Time to start playing with theme()!
#2 - Add text hierarchy
Time to start playing with theme()!
#2 - Add text hierarchy
Time to start playing with theme()!
#2 - Add text hierarchy
Time to start playing with theme()!
#2 - Add text hierarchy
Move away from the default fonts
#2 - Add text hierarchy
Move away from the default fonts
basic_plot +
theme(legend.position = "none",
text = element_text(colour = vit_c_palette["light_text"],
family = "Cabin"),
plot.title = element_text(colour = vit_c_palette["dark_text"],
size = rel(1.5),
face = "bold",
family = "Enriqueta"),
strip.text = element_text(family = "Enriqueta",
colour = vit_c_palette["light_text"],
size = rel(1.1), face = "bold"),
axis.text = element_text(colour = vit_c_palette["light_text"]))#2 - Add text hierarchy
#2 - Add text hierarchy
Choosing fonts can be tricky!
- Brand guidelines
- Datawrapper guidance - avoid fonts that are too wide/narrow!
- Websites + inspector tool
- Oliver Schöndorfer’s exploration of the Font Matrix
Find out more: pimpmytype.com/font-matrix/
#2 - Add text hierarchy
Getting custom fonts to work can be frustrating!
Install fonts locally, restart R Studio + 📦
{systemfonts}({ragg}+{textshaping}) + Set graphics device to “AGG” + 🤞
#2 - Add text hierarchy
See what fonts are available on your device
systemfonts::system_fonts() |> View()
#2 - Add text hierarchy
- Set of fonts + rules
- Set of colours + rules
A Dataviz Design System: A simple set of rules to follow, to effortlessly make your visualisations look on brand, every time you go to create a new plot.
- Accessibility
- Meaningful colours within your context
- “Visual identity”
#2 - Add text hierarchy
Give everything some space to breathe
basic_plot +
theme(legend.position = "none",
text = element_text(colour = vit_c_palette["light_text"],
family = "Cabin"),
plot.title = element_text(colour = vit_c_palette["dark_text"],
size = rel(1.5),
face = "bold",
family = "Enriqueta"),
strip.text = element_text(family = "Enriqueta",
colour = vit_c_palette["light_text"],
size = rel(1.1), face = "bold"),
axis.text = element_text(colour = vit_c_palette["light_text"]))#2 - Add text hierarchy
Give everything some space to breathe
basic_plot +
theme(legend.position = "none",
text = element_text(colour = vit_c_palette["light_text"],
family = "Cabin"),
plot.title = element_text(colour = vit_c_palette["dark_text"],
size = rel(1.5),
face = "bold",
family = "Enriqueta",
lineheight = 1.3,
margin = margin(0.5, 0, 1, 0, "lines")),
plot.subtitle = element_text(size = rel(1.1), lineheight = 1.3,
margin = margin(0, 0, 1, 0, "lines")),
strip.text = element_text(family = "Enriqueta",
colour = vit_c_palette["light_text"],
size = rel(1.1), face = "bold",
margin = margin(2, 0, 0.5, 0, "lines")),
axis.text = element_text(colour = vit_c_palette["light_text"]))#2 - Add text hierarchy
Remove unnecessary text
basic_plot +
theme(legend.position = "none",
text = element_text(colour = vit_c_palette["light_text"],
family = "Cabin"),
axis.title.y = element_blank(),
plot.title = element_text(colour = vit_c_palette["dark_text"],
size = rel(1.5),
face = "bold",
family = "Enriqueta",
lineheight = 1.3,
margin = margin(0.5, 0, 1, 0, "lines")),
plot.subtitle = element_text(size = rel(1.1), lineheight = 1.3,
margin = margin(0, 0, 1, 0, "lines")),
strip.text = element_text(family = "Enriqueta",
colour = vit_c_palette["light_text"],
size = rel(1.1), face = "bold",
margin = margin(2, 0, 0.5, 0, "lines")),
axis.text = element_text(colour = vit_c_palette["light_text"]))#2 - Add text hierarchy
Watch out for that title!
basic_plot +
labs(title = "In smaller doses, Orange Juice was associated with greater mean tooth growth,
compared to equivalent doses of Vitamin C") +
theme(legend.position = "none",
text = element_text(colour = vit_c_palette["light_text"],
family = "Cabin"),
axis.title.y = element_blank(),
plot.title = element_text(colour = vit_c_palette["dark_text"],
size = 36,
face = "bold",
family = "Enriqueta",
lineheight = 1.3,
margin = margin(0.5, 0, 1, 0, "lines")),
plot.subtitle = element_text(size = rel(1.1), lineheight = 1.3,
margin = margin(0, 0, 1, 0, "lines")),
strip.text = element_text(family = "Enriqueta",
colour = vit_c_palette["light_text"],
size = rel(1.1), face = "bold",
margin = margin(2, 0, 0.5, 0, "lines")),
axis.text = element_text(colour = vit_c_palette["light_text"]))#2 - Add text hierarchy
Watch out for that title!
basic_plot +
labs(title = "In smaller doses, Orange Juice was associated with greater mean tooth growth, compared to equivalent doses of Vitamin C") +
theme(legend.position = "none",
text = element_text(colour = vit_c_palette["light_text"],
family = "Cabin"),
axis.title.y = element_blank(),
plot.title = element_text(colour = vit_c_palette["dark_text"],
size = rel(1.5),
face = "bold",
family = "Enriqueta",
lineheight = 1.3,
margin = margin(0.5, 0, 1, 0, "lines")),
plot.subtitle = element_text(size = rel(1.1), lineheight = 1.3,
margin = margin(0, 0, 1, 0, "lines")),
strip.text = element_text(family = "Enriqueta",
colour = vit_c_palette["light_text"],
size = rel(1.1), face = "bold",
margin = margin(2, 0, 0.5, 0, "lines")),
axis.text = element_text(colour = vit_c_palette["light_text"]))#2 - Add text hierarchy
I ❤️ 📦 {ggtext}
basic_plot +
labs(title = "In smaller doses, Orange Juice was associated with greater mean tooth growth, compared to equivalent doses of Vitamin C") +
theme(legend.position = "none",
text = element_text(colour = vit_c_palette["light_text"],
family = "Cabin"),
axis.title.y = element_blank(),
plot.title = ggtext::element_textbox_simple(
colour = vit_c_palette["dark_text"],
size = rel(1.5),
face = "bold",
family = "Enriqueta",
lineheight = 1.3,
margin = margin(0.5, 0, 1, 0, "lines")),
plot.subtitle = ggtext::element_textbox_simple(
size = rel(1.1),
lineheight = 1.3,
margin = margin(0, 0, 1, 0, "lines")),
strip.text = element_text(family = "Enriqueta",
colour = vit_c_palette["light_text"],
size = rel(1.1), face = "bold",
margin = margin(2, 0, 0.5, 0, "lines")),
axis.text = element_text(colour = vit_c_palette["light_text"]))#2 - Add text hierarchy + colour!
Hello, HTML + CSS!
We can make text <span style='color:green'>green</span> and also <span style='color:green; font-size:60pt'>really big</span>! 🤯
We can make text green and also really big! 🤯
#2 - Add text hierarchy + colour!
I ❤️ 📦 {ggtext}
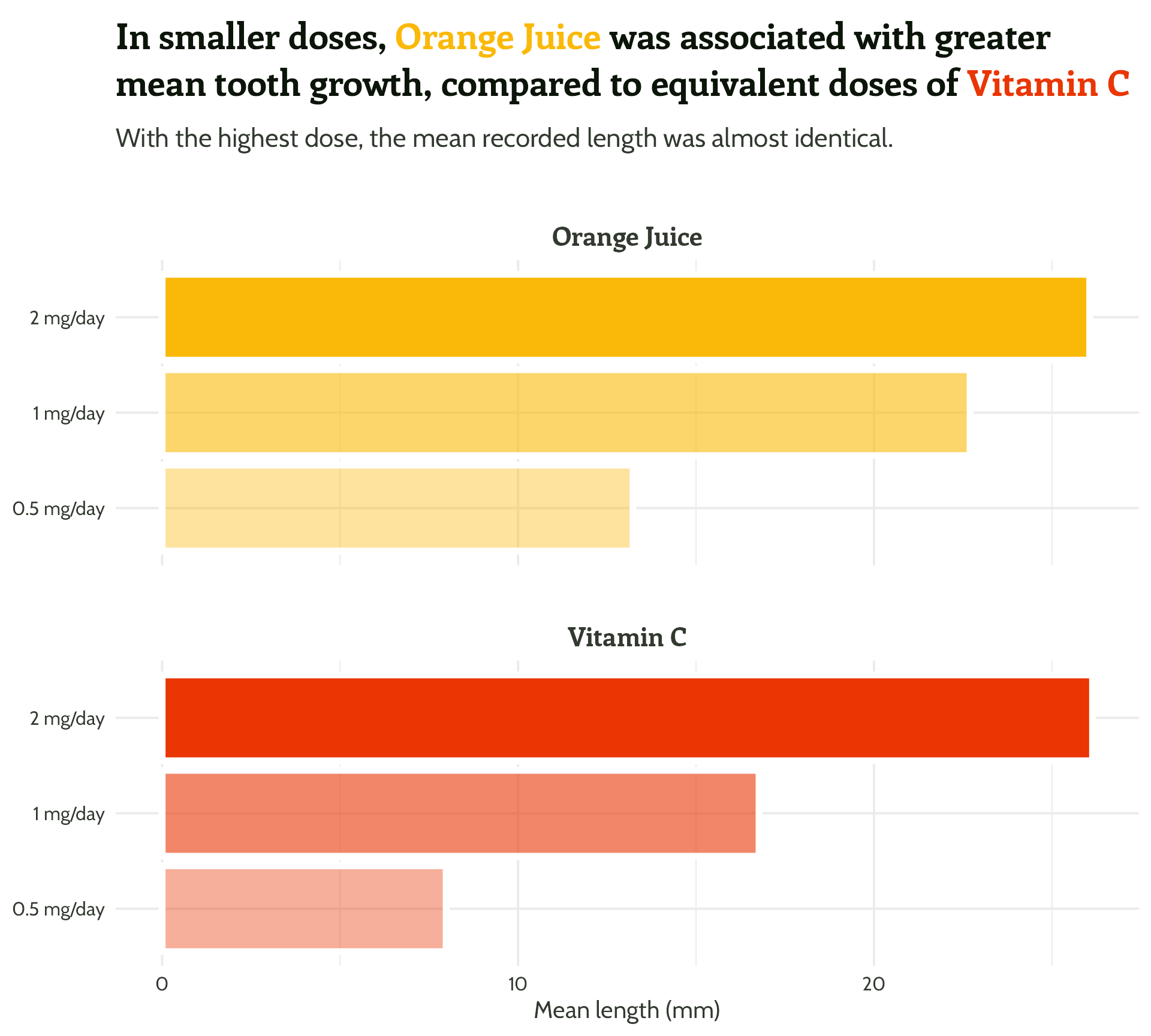
basic_plot +
labs(title =
paste0("In smaller doses, **<span style='color:",
vit_c_palette["Orange Juice"], "'>Orange Juice</span>**
was associated with greater mean tooth growth,
compared to equivalent doses of **<span style='color:",
vit_c_palette["Vitamin C"], "'>Vitamin C</span>**")
) +
theme(legend.position = "none",
text = element_text(colour = vit_c_palette["light_text"],
family = "Cabin"),
axis.title.y = element_blank(),
plot.title = ggtext::element_textbox_simple(colour = vit_c_palette["dark_text"],
size = rel(1.5),
face = "bold",
family = "Enriqueta",
lineheight = 1.3,
margin = margin(0.5, 0, 1, 0, "lines")),
plot.subtitle = ggtext::element_textbox_simple(family = "Cabin", size = rel(1.1), lineheight = 1.3,
margin = margin(0, 0, 1, 0, "lines")),
strip.text = element_text(family = "Enriqueta",
colour = vit_c_palette["light_text"],
size = rel(1.1), face = "bold",
margin = margin(2, 0, 0.5, 0, "lines")),
axis.text = element_text(colour = vit_c_palette["light_text"]))#2 - Add text hierarchy + colour!
#2 - Add text hierarchy
See for yourselves!
Over to you!
- See what fonts are available on your device -
systemfonts::systemfonts() |> View() - If you want to, install a new one!
- Apply one to the title, and one to the rest of the text using
theme() - Create some text colour variants using
monochromeR::generate_palette() - Apply those to the title and the other text elements
- Play around with text size
See you in 10 minutes! 📊 🎨 ☕
10:00
Package up!
At this point, we’re on our way to implementing our Dataviz Design System in R. Let’s package up a few things for use in future plots!
Packaging up
- Package development is a whole other workshop (but it’s easier than you think!)
- 📦
{usethis}
- 📦
- Any function or object you create can be added to a package
green blue dark_text light_text
"#28A569" "#2C3D4F" "#1A242F" "#808A95" Find out more: Shannon Pileggi - Your first R package in 1 hour
Packaging up
Packaging up
theme_guineapigs <- function(base_text_size = 15,
palette = vit_c_palette) {
theme_minimal(base_size = base_text_size) +
theme(legend.position = "none",
text = element_text(colour = palette["light_text"],
family = "Cabin"),
axis.title.y = element_blank(),
plot.title = ggtext::element_textbox_simple(colour = palette["dark_text"],
size = rel(1.5),
face = "bold",
family = "Enriqueta",
lineheight = 1.3,
margin = margin(0.5, 0, 1, 0, "lines")),
plot.subtitle = ggtext::element_textbox_simple(family = "Cabin", size = rel(1.1), lineheight = 1.3,
margin = margin(0, 0, 1, 0, "lines")),
strip.text = element_text(family = "Enriqueta",
colour = palette["light_text"],
size = rel(1.1), face = "bold",
margin = margin(2, 0, 0.5, 0, "lines")),
axis.text = element_text(colour = palette["light_text"]))
}Packaging up
Packaging up
Five tips for beautiful annotations
#1 - Use colour purposefully
#2 - Add text hierarchy
#3 - Reduce unnecessary eye movement
#4 - Highlight important patterns
#5 - See how much you can declutter
Bonus track - {gghighlight} & curved arrows
#3 - Reduce unnecessary eye movement
We’ve made it easy to see what’s what. Now, let’s make it even easier to compare values.
#3 - Reduce unnecessary eye movement
We’ve made it easy to see what’s what. Now, let’s make it even easier to compare values.
#3 - Reduce unnecessary eye movement
We’ve made it easy to see what’s what. Now, let’s make it even easier to compare values.
#3 - Reduce unnecessary eye movement
Time to add some text boxes!
#3 - Reduce unnecessary eye movement
Time to add some text boxes!
Find out more: Alignment Cheatsheet
#3 - Reduce unnecessary eye movement
Time to add some text boxes!
#3 - Reduce unnecessary eye movement
Time to add some text boxes!
#3 - Reduce unnecessary eye movement
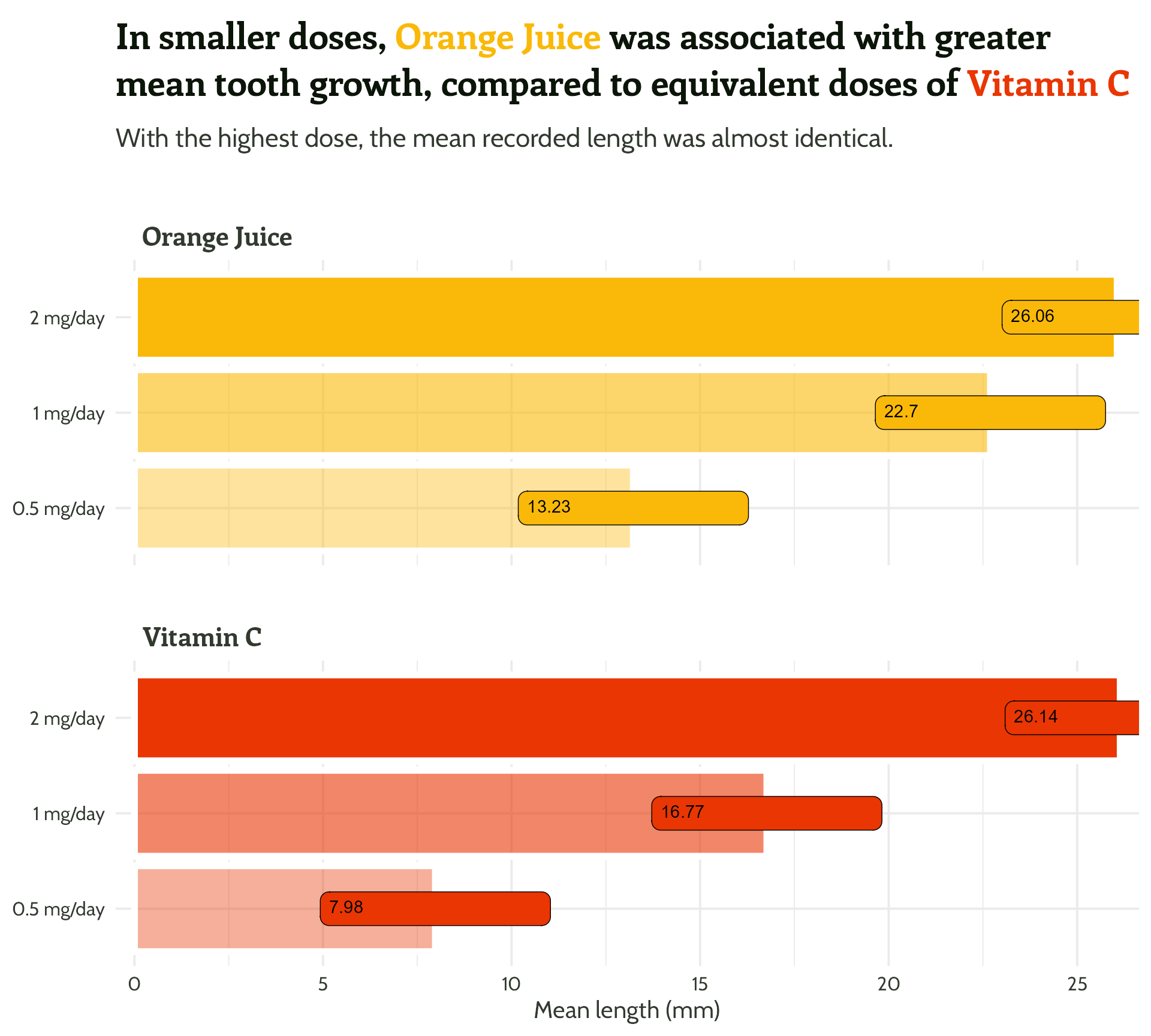
Now for the fun stuff…
themed_plot +
scale_y_continuous(expand = c(0, 0.5)) +
theme(strip.text = element_text(hjust = 0.03)) +
ggtext::geom_textbox(aes(
label = mean_length,
hjust = case_when(mean_length < 15 ~ 0,
TRUE ~ 1),
halign = case_when(mean_length < 15 ~ 0,
TRUE ~ 1)),
size = 6,
fill = NA,
box.colour = NA,
family = "Cabin",
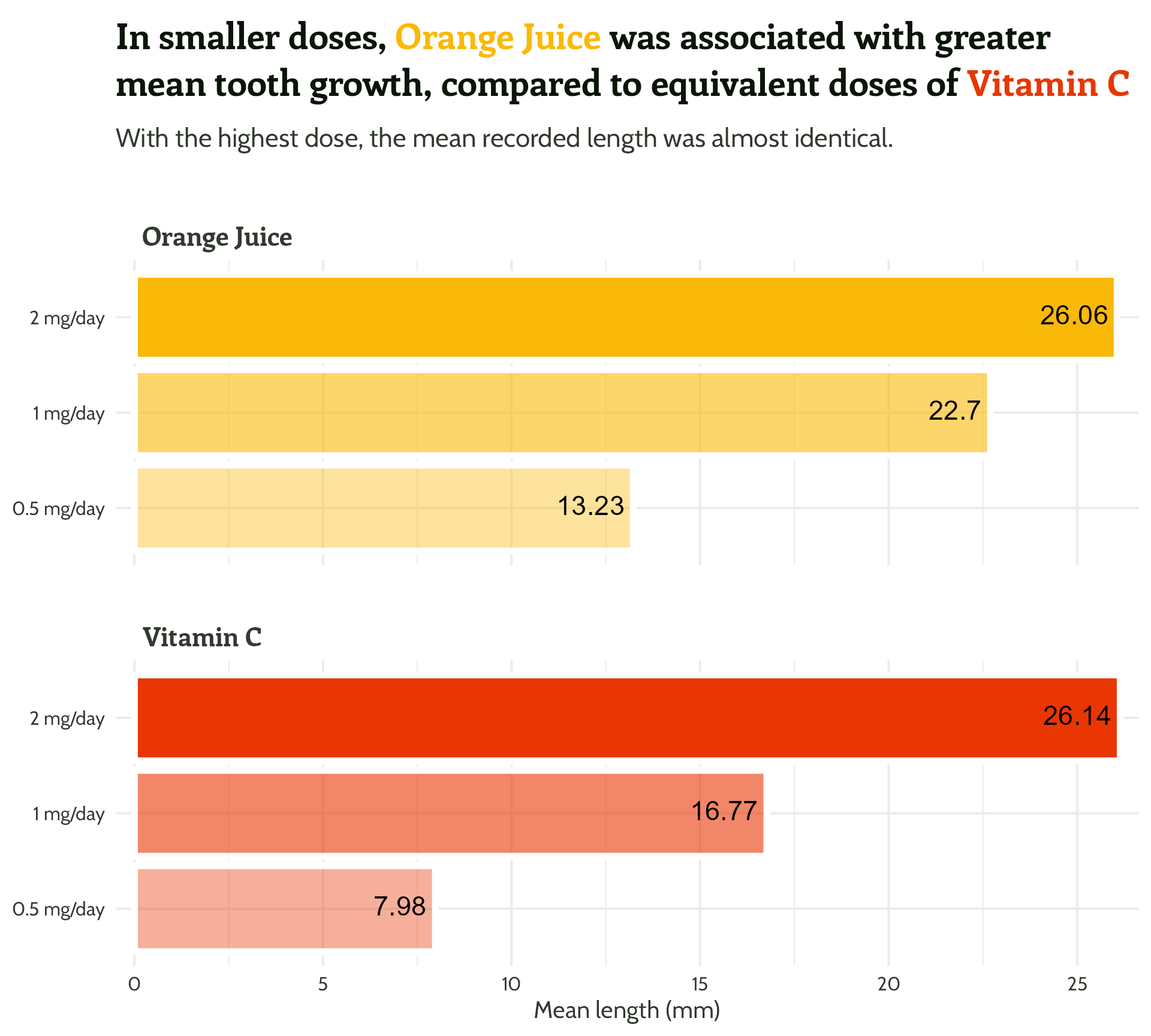
fontface = "bold")#3 - Reduce unnecessary eye movement
Now for the fun stuff…
themed_plot +
scale_y_continuous(expand = c(0, 0.5)) +
theme(strip.text = element_text(hjust = 0.03)) +
ggtext::geom_textbox(aes(
label = mean_length,
hjust = case_when(mean_length < 15 ~ 0,
TRUE ~ 1),
halign = case_when(mean_length < 15 ~ 0,
TRUE ~ 1),
colour = case_when(mean_length > 15 ~ "#FFFFFF",
TRUE ~ vit_c_palette[supplement])),
size = 6,
fill = NA,
box.colour = NA,
family = "Cabin",
fontface = "bold")#3 - Reduce unnecessary eye movement
??????
themed_plot +
scale_y_continuous(expand = c(0, 0.5)) +
theme(strip.text = element_text(hjust = 0.03)) +
ggtext::geom_textbox(aes(
label = mean_length,
hjust = case_when(mean_length < 15 ~ 0,
TRUE ~ 1),
halign = case_when(mean_length < 15 ~ 0,
TRUE ~ 1),
colour = case_when(mean_length > 15 ~ "#FFFFFF",
TRUE ~ vit_c_palette[supplement])),
size = 6,
fill = NA,
box.colour = NA,
family = "Cabin",
fontface = "bold")#3 - Reduce unnecessary eye movement
scale_colour_identity() required!
themed_plot +
scale_y_continuous(expand = c(0, 0.5)) +
theme(strip.text = element_text(hjust = 0.03)) +
scale_colour_identity() +
ggtext::geom_textbox(aes(
label = mean_length,
hjust = case_when(mean_length < 15 ~ 0,
TRUE ~ 1),
halign = case_when(mean_length < 15 ~ 0,
TRUE ~ 1),
colour = case_when(mean_length > 15 ~ "#FFFFFF",
TRUE ~ vit_c_palette[supplement])),
size = 6,
fill = NA,
box.colour = NA,
family = "Cabin",
fontface = "bold")#3 - Reduce unnecessary eye movement
We might as well add a bit of extra info (with text hierarchy!) to our labels…
themed_plot +
scale_y_continuous(expand = c(0, 0.5)) +
theme(strip.text = element_text(hjust = 0.03)) +
scale_colour_identity() +
ggtext::geom_textbox(aes(
label = paste0("<span style=font-size:12pt>",
dose, "mg/day</span><br>",
mean_length, "mm"),
hjust = case_when(mean_length < 15 ~ 0,
TRUE ~ 1),
halign = case_when(mean_length < 15 ~ 0,
TRUE ~ 1),
colour = case_when(mean_length > 15 ~ "#FFFFFF",
TRUE ~ vit_c_palette[supplement])),
size = 6,
fill = NA,
box.colour = NA,
family = "Cabin",
fontface = "bold")Wait, but why?
#3 - Reduce unnecessary eye movement
#3 - Reduce unnecessary eye movement
Easier than you think and makes a big difference! 🦸
Over to you!
- Add a textbox (or several!) using
ggtext::geom_textbox() - Add in some styling and conditional alignments
- See what happens when you apply your plot code to random subsets of your data
See you in 5 minutes! 📊 🎨 ☕
05:00
Five tips for beautiful annotations
#1 - Use colour purposefully
#2 - Add text hierarchy
#3 - Reduce unnecessary eye movement
#4 - Highlight important patterns
#5 - See how much you can declutter
Bonus track - {gghighlight}, Tee pipe and curved arrows
#4 - Highlight important patterns
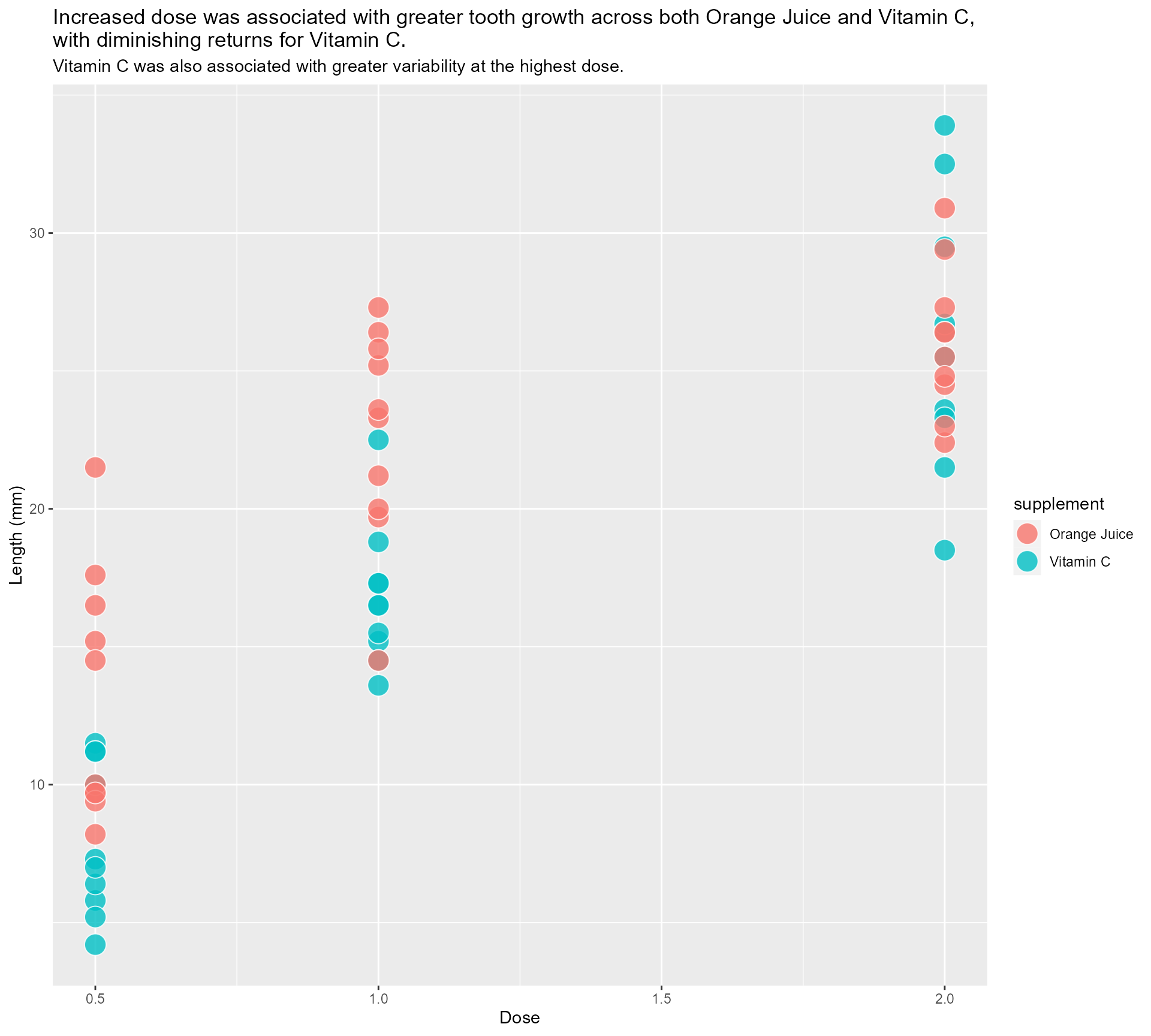
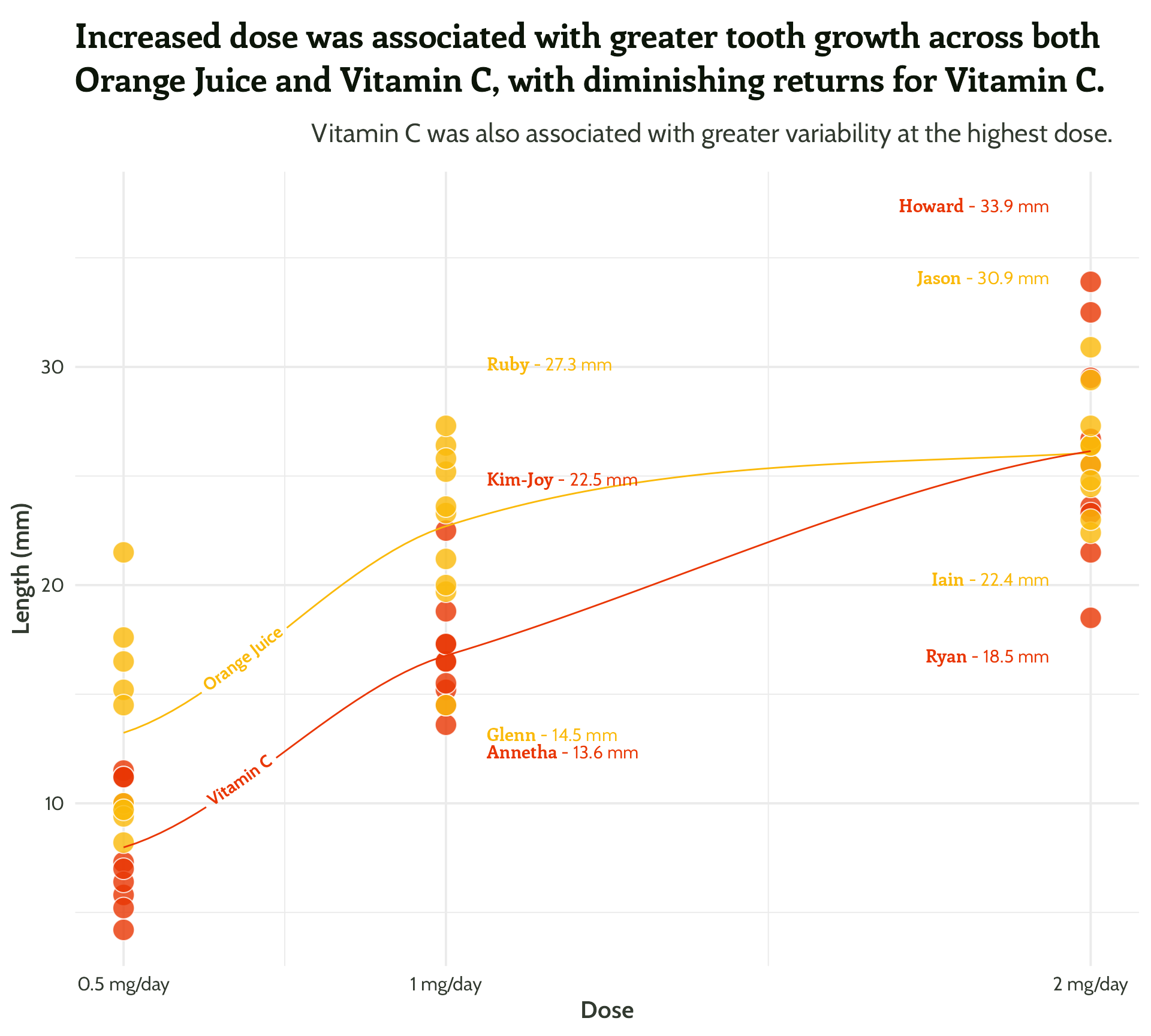
“That’s all well and good, but we all know summary data can be misleading…”
Find out more: cararthompson.com/talks/nhsr2022-ggplot-themes
#4 - Highlight important patterns
I ❤️ 📦 {geomtextpath}
#4 - Highlight important patterns
I ❤️ 📦 {geomtextpath}
#4 - Highlight important patterns
I ❤️ 📦 {geomtextpath}
#4 - Highlight important patterns
I ❤️ 📦 {geomtextpath}
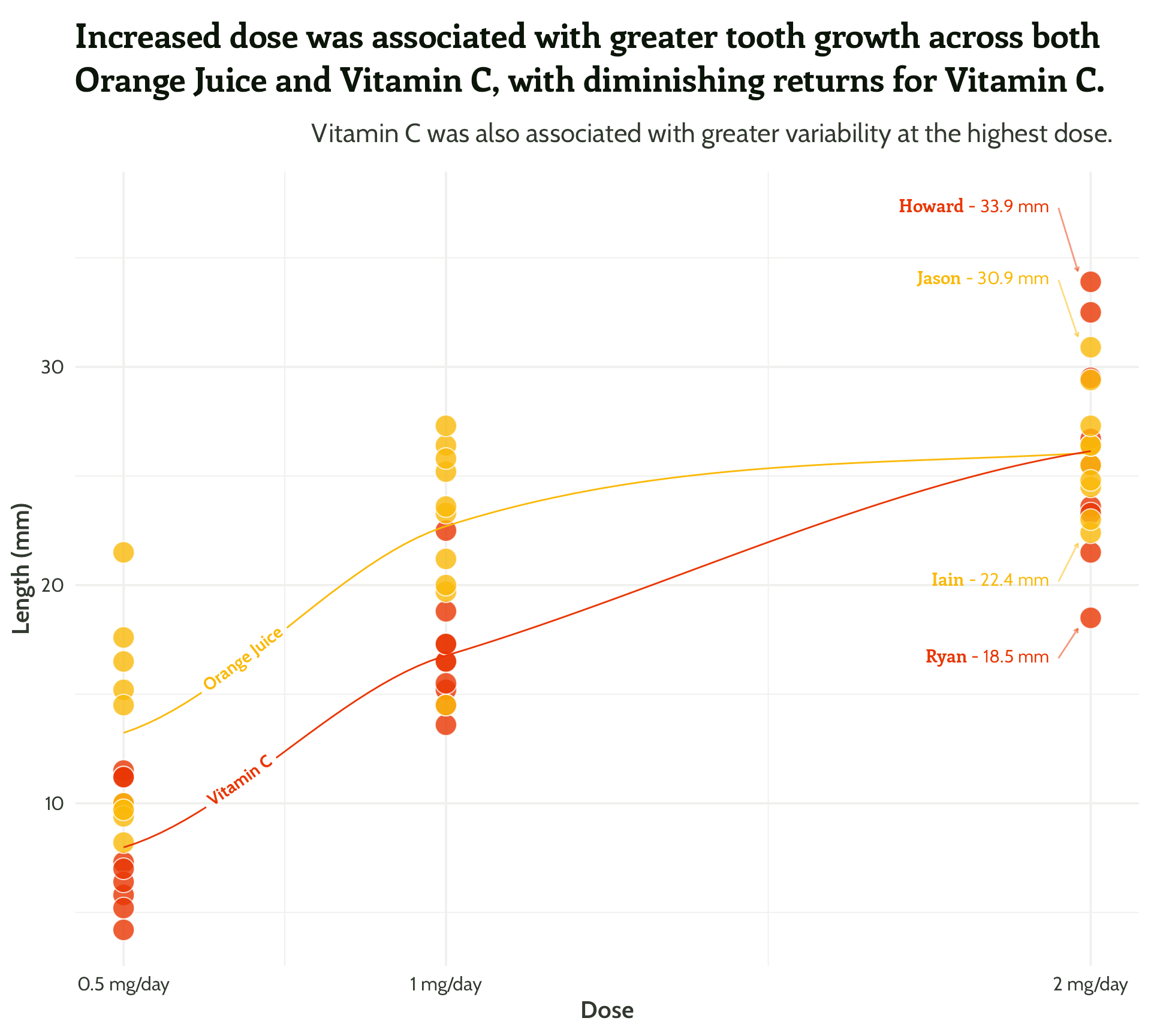
#4 - Highlight important patterns
More textboxes with markdown and conditional alignment (horizontal and vertical!)
themed_scatter_plot +
geomtextpath::geom_textline(stat = "smooth", aes(label = supplement),
hjust = 0.1,
vjust = 0.3,
fontface = "bold",
family = "Cabin") +
ggtext::geom_textbox(data = filter(min_max_gps,
dose %in% c(1, 2)),
aes(x = case_when(dose < 1.5 ~ dose + 0.05,
TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1,
TRUE ~ len * 0.9),
label = paste0("**<span style='font-family:Enriqueta'>",
guinea_pig_name,
"</span>** - ", len, " mm"),
hjust = case_when(dose < 1.5 ~ 0,
TRUE ~ 1),
halign = case_when(dose < 1.5 ~ 0,
TRUE ~ 1)),
family = "Cabin",
size = 4,
fill = NA,
box.colour = NA) +
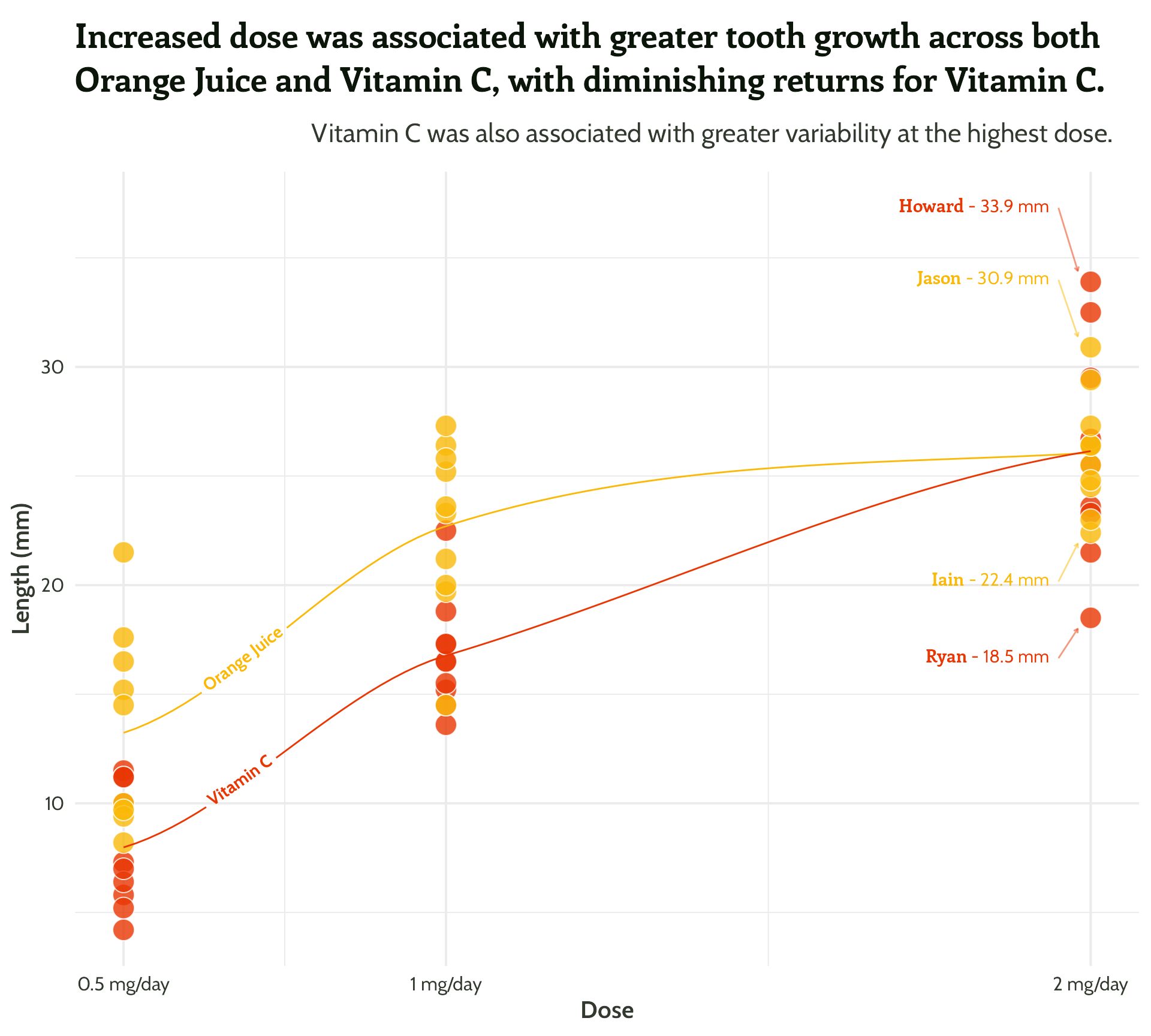
scale_colour_manual(values = vit_c_palette)#4 - Highlight important patterns
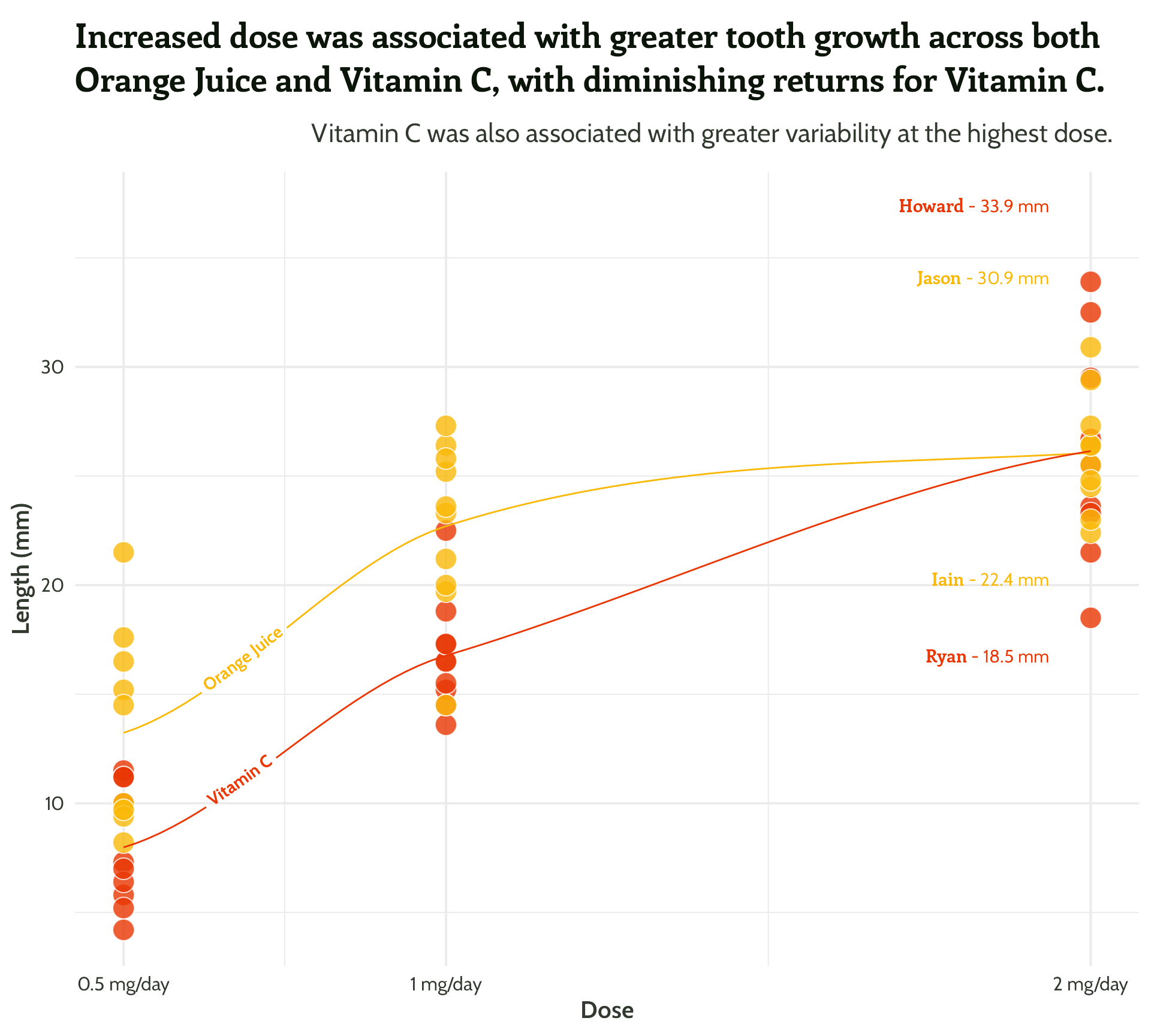
Sometimes less is more!
themed_scatter_plot +
geomtextpath::geom_textline(stat = "smooth", aes(label = supplement),
hjust = 0.1,
vjust = 0.3,
fontface = "bold",
family = "Cabin") +
ggtext::geom_textbox(data = filter(min_max_gps,
dose == 2),
aes(x = case_when(dose < 1.5 ~ dose + 0.05,
TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1,
TRUE ~ len * 0.9),
label = paste0("**<span style='font-family:Enriqueta'>",
guinea_pig_name,
"</span>** - ", len, " mm"),
hjust = case_when(dose < 1.5 ~ 0,
TRUE ~ 1),
halign = case_when(dose < 1.5 ~ 0,
TRUE ~ 1)),
family = "Cabin",
size = 4,
fill = NA,
box.colour = NA) +
scale_colour_manual(values = vit_c_palette)#4 - Highlight important patterns
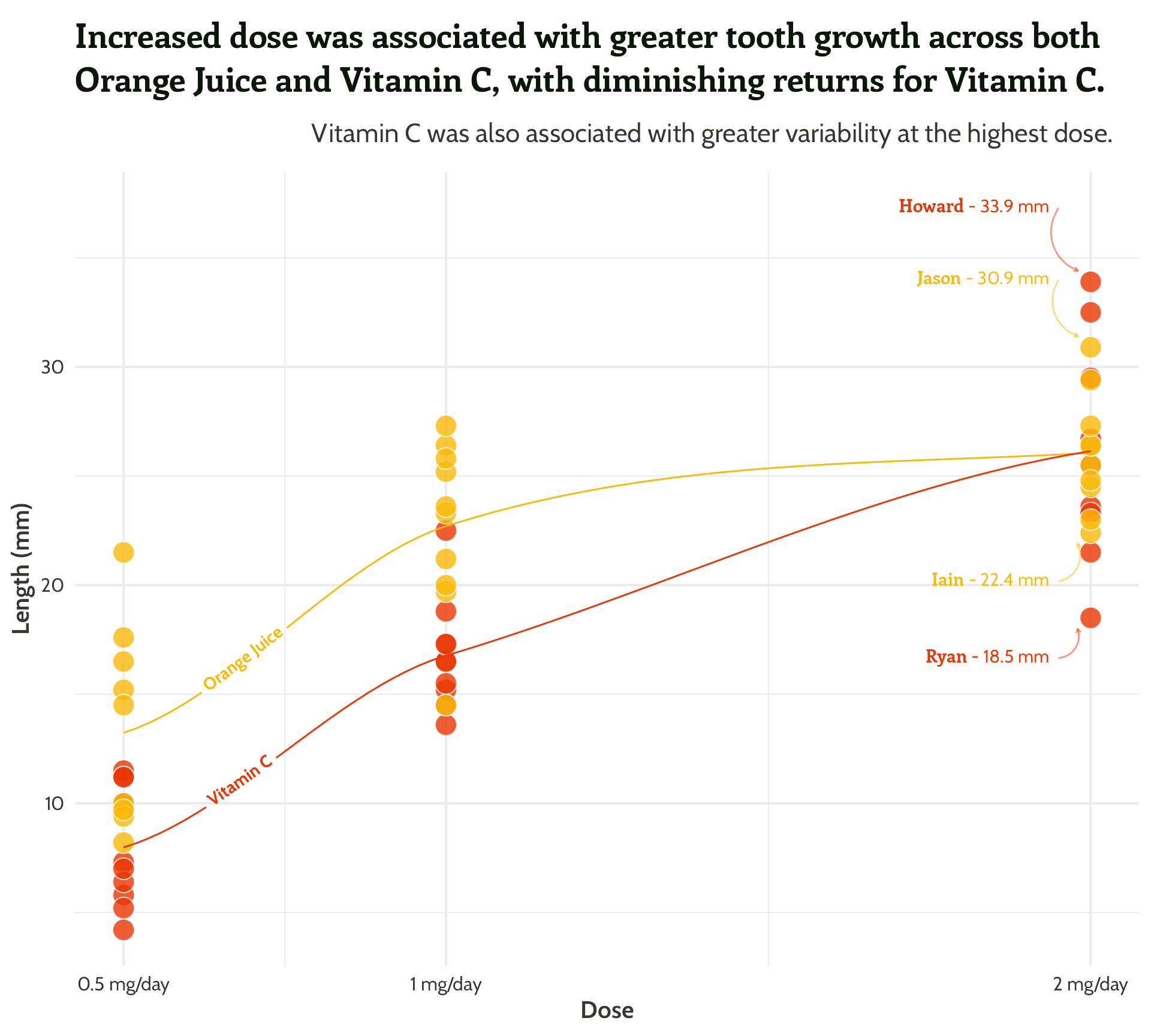
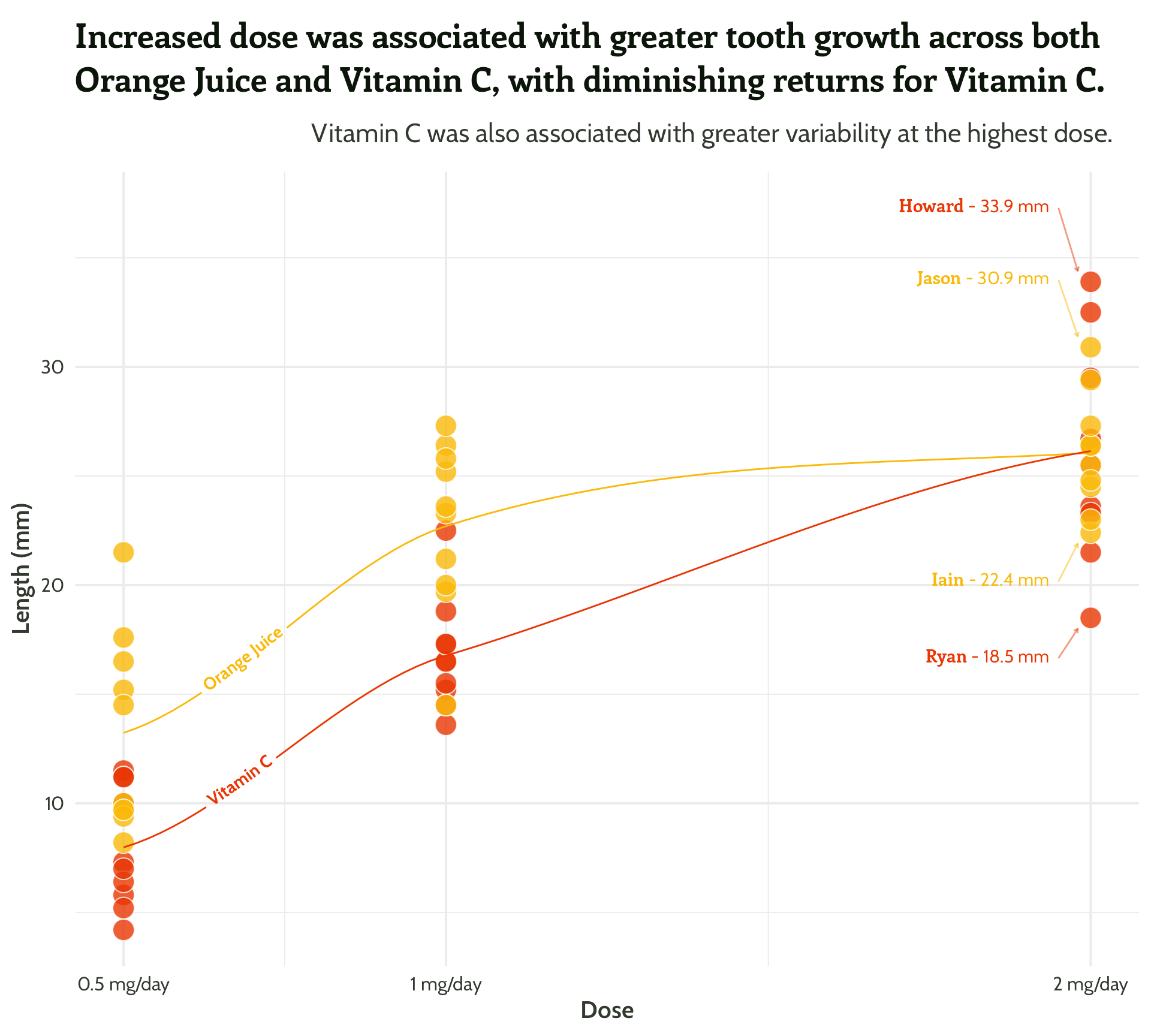
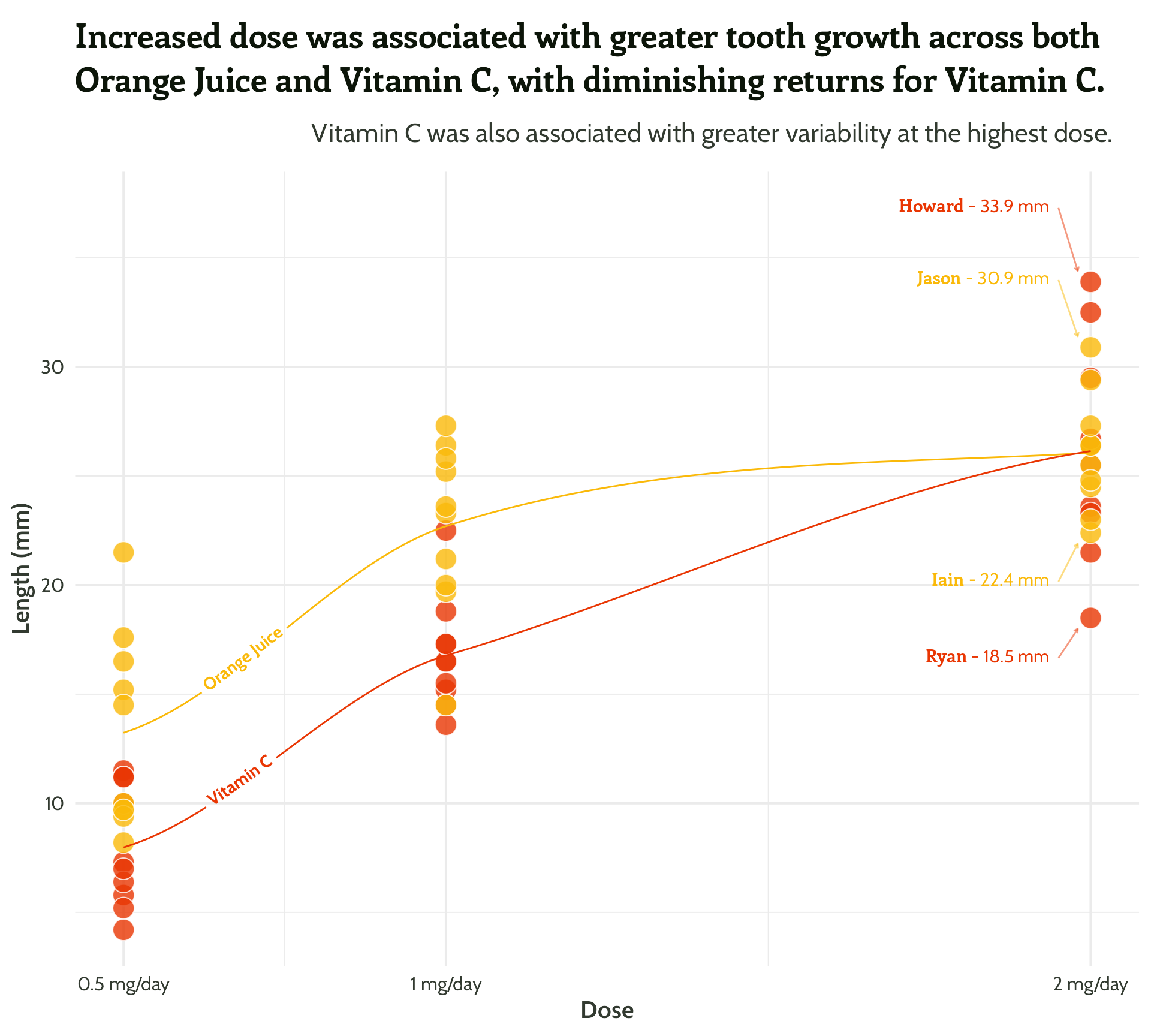
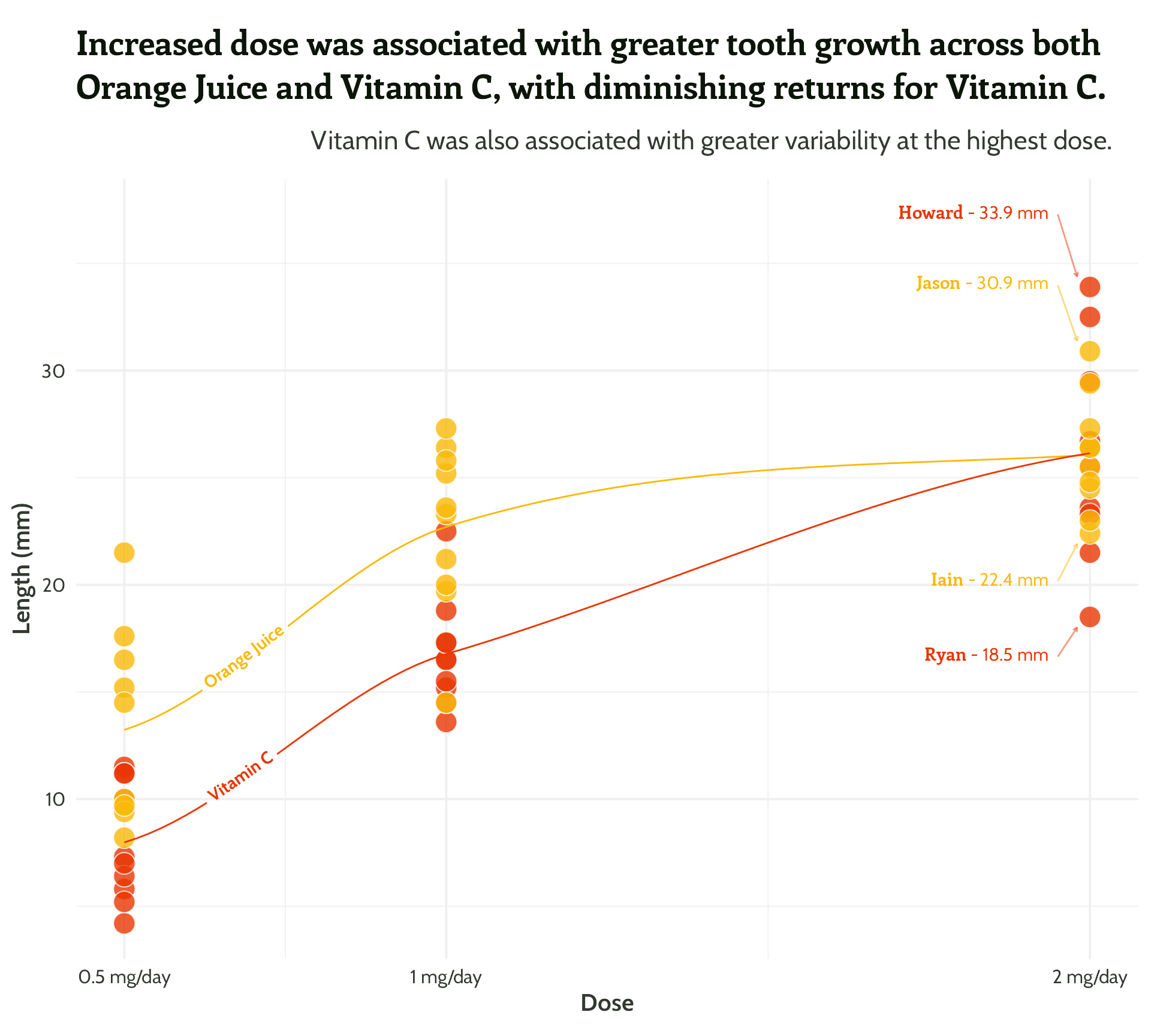
Same principle, let’s add in some arrows!
themed_scatter_plot +
geomtextpath::geom_textline(stat = "smooth", aes(label = supplement),
hjust = 0.1,
vjust = 0.3,
fontface = "bold",
family = "Cabin") +
ggtext::geom_textbox(data = filter(min_max_gps,
dose == 2),
aes(x = case_when(dose < 1.5 ~ dose + 0.05, TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1, TRUE ~ len * 0.9),
label = paste0("**<span style='font-family:Enriqueta'>", guinea_pig_name,"</span>** - ", len, " mm"),
hjust = case_when(dose < 1.5 ~ 0,TRUE ~ 1),
halign = case_when(dose < 1.5 ~ 0, TRUE ~ 1)),
family = "Cabin", size = 4, fill = NA, box.colour = NA) +
geom_curve(data = filter(min_max_gps,
dose == 2),
aes(x = case_when(dose < 1.5 ~ dose + 0.05,
TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1,
TRUE ~ len * 0.9),
xend = case_when(dose < 1.5 ~ dose + 0.02,
TRUE ~ dose - 0.02),
yend = case_when(min_or_max == "max"~ len + 0.5,
TRUE ~ len - 0.5)),
arrow = arrow(length = unit(0.1, "cm")),
alpha = 0.5) +
scale_colour_manual(values = vit_c_palette)#4 - Highlight important patterns
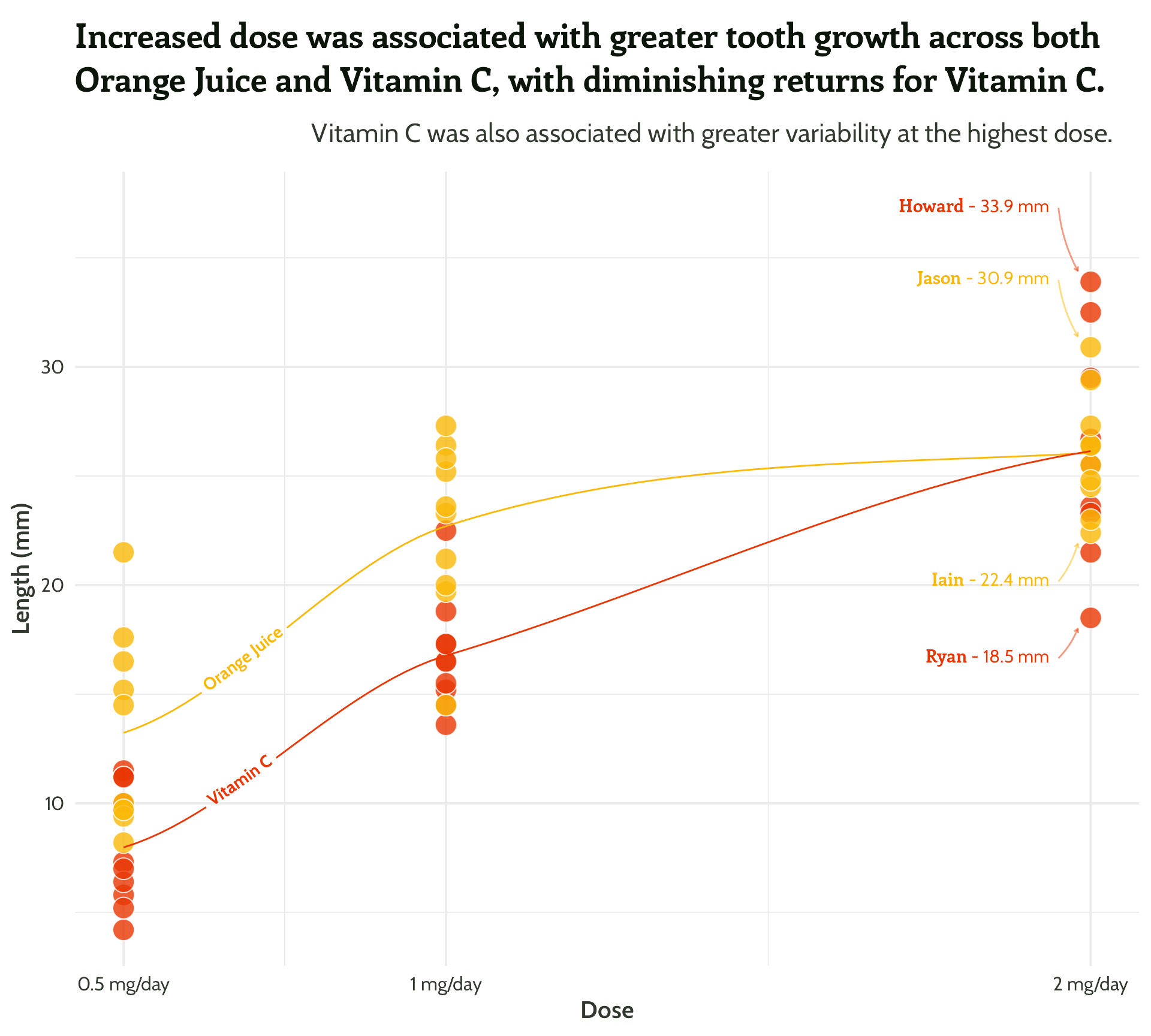
Same principle, let’s add in some arrows!
themed_scatter_plot +
geomtextpath::geom_textline(stat = "smooth", aes(label = supplement),
hjust = 0.1,
vjust = 0.3,
fontface = "bold",
family = "Cabin") +
ggtext::geom_textbox(data = filter(min_max_gps,
dose == 2),
aes(x = case_when(dose < 1.5 ~ dose + 0.05, TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1, TRUE ~ len * 0.9),
label = paste0("**<span style='font-family:Enriqueta'>", guinea_pig_name,"</span>** - ", len, " mm"),
hjust = case_when(dose < 1.5 ~ 0,TRUE ~ 1),
halign = case_when(dose < 1.5 ~ 0, TRUE ~ 1)),
family = "Cabin", size = 4, fill = NA, box.colour = NA) +
geom_curve(data = filter(min_max_gps,
dose == 2),
aes(x = case_when(dose < 1.5 ~ dose + 0.05,
TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1,
TRUE ~ len * 0.9),
xend = case_when(dose < 1.5 ~ dose + 0.02,
TRUE ~ dose - 0.02),
yend = case_when(min_or_max == "max"~ len + 0.5,
TRUE ~ len - 0.5)),
curvature = 0.1,
arrow = arrow(length = unit(0.1, "cm")),
alpha = 0.5) +
scale_colour_manual(values = vit_c_palette)#4 - Highlight important patterns
Same principle, let’s add in some arrows!
themed_scatter_plot +
geomtextpath::geom_textline(stat = "smooth", aes(label = supplement),
hjust = 0.1,
vjust = 0.3,
fontface = "bold",
family = "Cabin") +
ggtext::geom_textbox(data = filter(min_max_gps,
dose == 2),
aes(x = case_when(dose < 1.5 ~ dose + 0.05, TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1, TRUE ~ len * 0.9),
label = paste0("**<span style='font-family:Enriqueta'>", guinea_pig_name,"</span>** - ", len, " mm"),
hjust = case_when(dose < 1.5 ~ 0,TRUE ~ 1),
halign = case_when(dose < 1.5 ~ 0, TRUE ~ 1)),
family = "Cabin", size = 4, fill = NA, box.colour = NA) +
geom_curve(data = filter(min_max_gps,
dose == 2),
aes(x = case_when(dose < 1.5 ~ dose + 0.05,
TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1,
TRUE ~ len * 0.9),
xend = case_when(dose < 1.5 ~ dose + 0.02,
TRUE ~ dose - 0.02),
yend = case_when(min_or_max == "max"~ len + 0.5,
TRUE ~ len - 0.5)),
curvature = 0,
arrow = arrow(length = unit(0.1, "cm")),
alpha = 0.5) +
scale_colour_manual(values = vit_c_palette)#4 - Highlight important patterns
Nearly there, folks! Look how far we’ve come!
#4 - Highlight important patterns
Five tips for beautiful annotations
#1 - Use colour purposefully
#2 - Add text hierarchy
#3 - Reduce unnecessary eye movement
#4 - Highlight important patterns
#5 - See how much you can declutter
Bonus track - {gghighlight}, Tee pipe and curved arrows
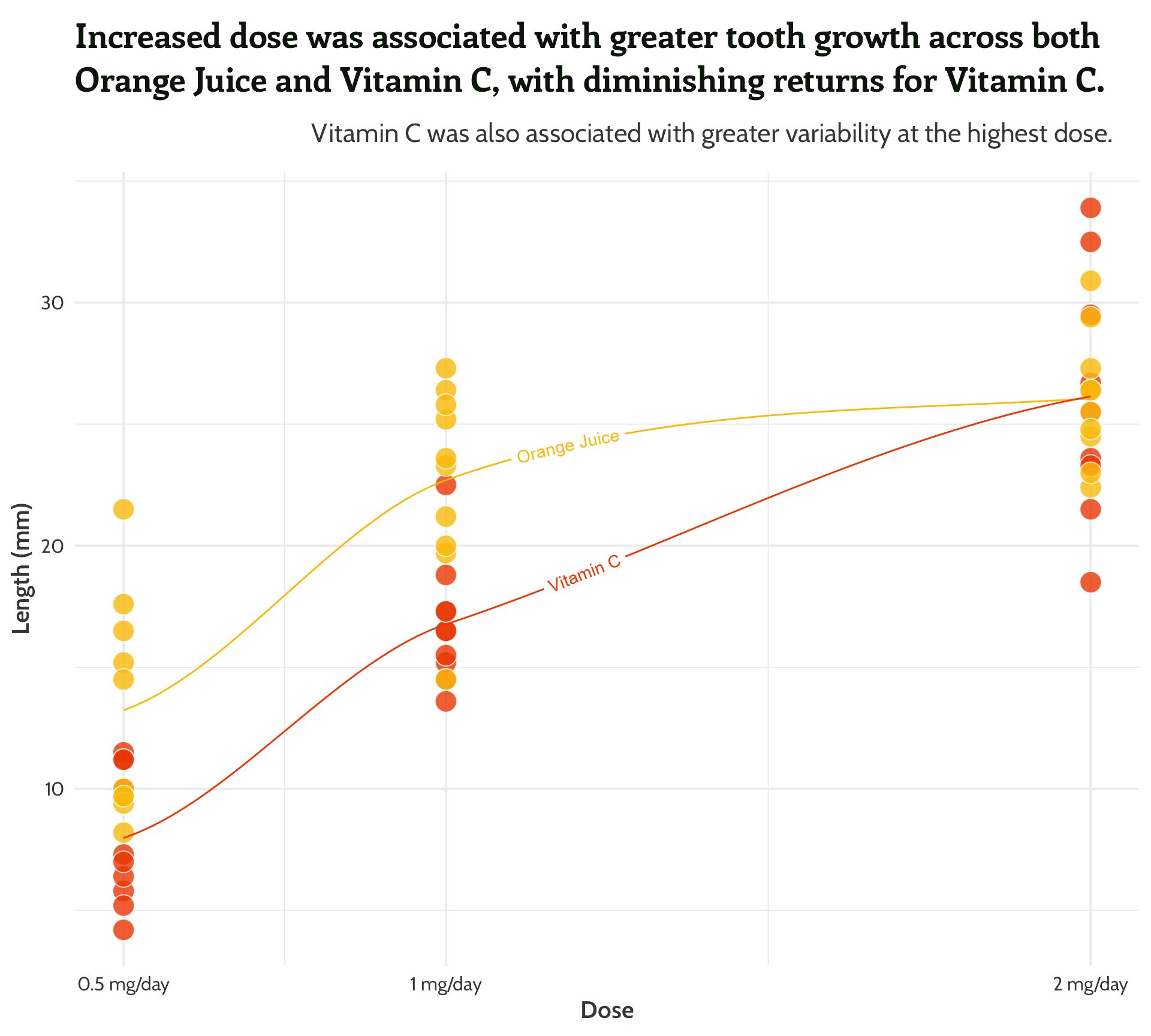
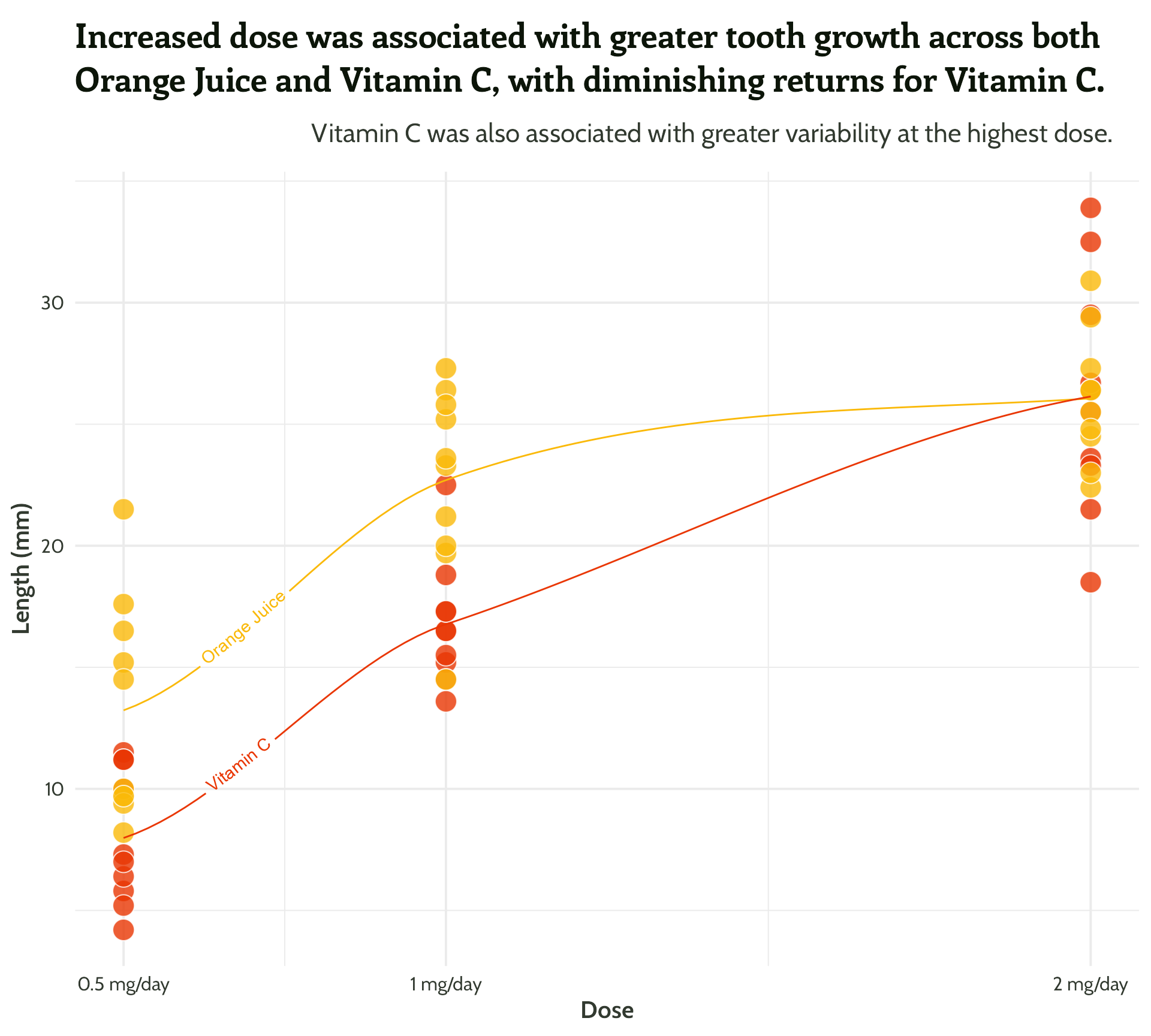
#5 - See how much you can declutter
#5 - See how much you can declutter
Tweak the grid lines, using a matching colour - {monochromeR}
#5 - See how much you can declutter
And add a bit of white space
#5 - See how much you can declutter
Five tips for beautiful annotations
#1 - Use colour purposefully
#2 - Add text hierarchy
#3 - Reduce unnecessary eye movement
#4 - Highlight important patterns
#5 - See how much you can declutter
Bonus track - {gghighlight}, Tee pipe and curved arrows
Bonus track - {gghighlight}, Tee pipe and curved arrows
{gghighlight}
“But I have way more conditions than this, how can I highlight a more subtle pattern?”
Suppose we have data that has so many series that it is hard to identify them by their colours as the differences are so subtle…
Find out more: yutannihilation.github.io/gghighlight/
{gghighlight}
Nicola Rennie’s Hollywood Age Gaps plot
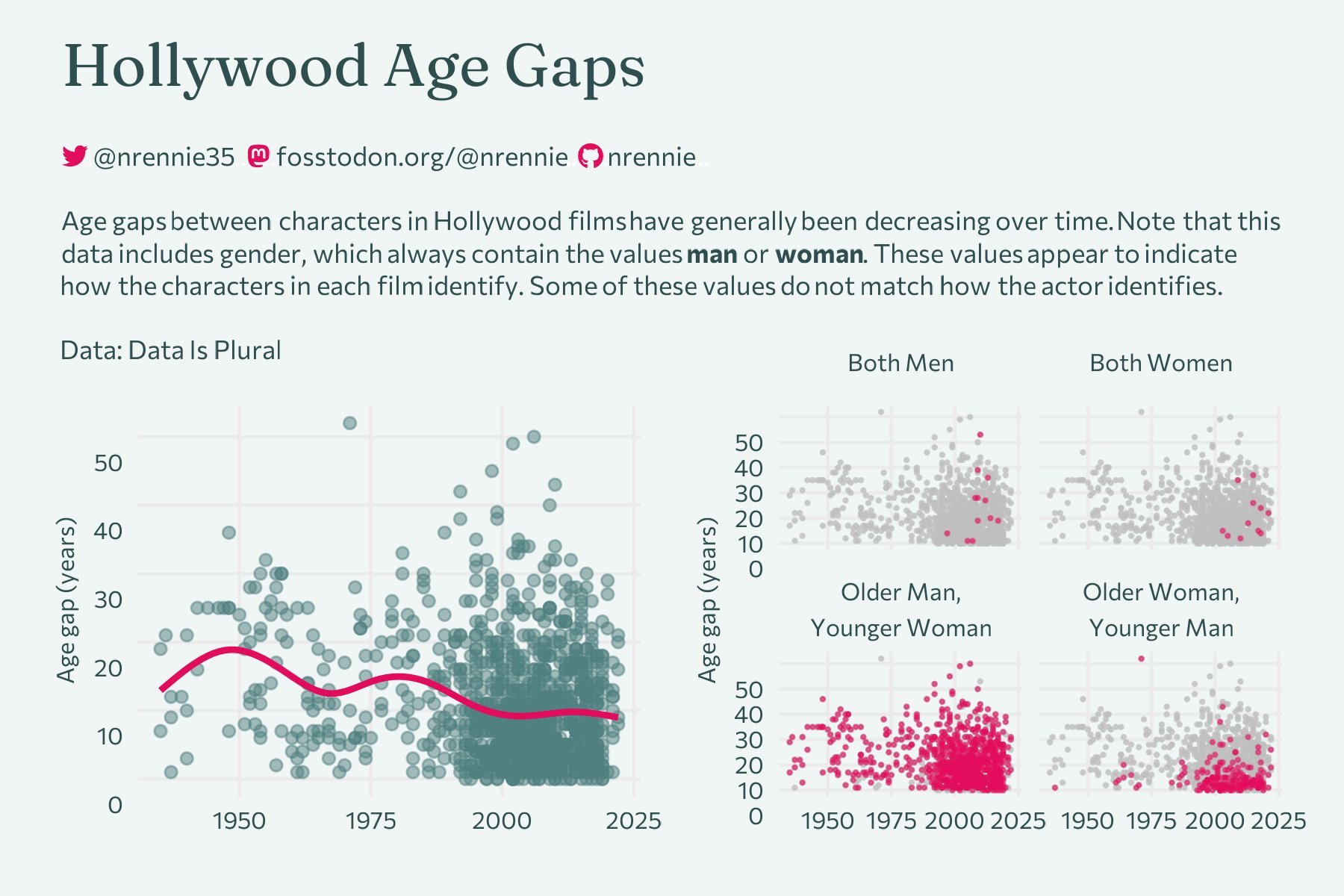
Find out more: twitter.com/nrennie35
Tee pipe
Creating label content on the fly
ToothGrowth %>%
mutate(guinea_pig_name = sample(unique(bakeoff::bakers$baker), 60),
supplement = case_when(supp == "OJ" ~ "Orange Juice",
supp == "VC" ~ "Vitamin C",
TRUE ~ as.character(supp))) %T>%
{
{
# Double assign to jump out of the pipe!
min_max_gps <<- group_by(., supplement, dose) %>%
filter(., len == min(len) | len == max(len)) %>%
mutate(min_or_max = case_when(len == max(len) ~ "max",
TRUE ~ "min"))
}
} %>%
ggplot(aes(x = dose, y = len, fill = supplement,
colour = supplement)) +
...Curved arrows
Conditional curvatures?
themed_scatter_plot +
geomtextpath::geom_textline(stat = "smooth", aes(label = supplement), hjust = 0.1, vjust = 0.3, fontface = "bold", family = "Cabin") +
ggtext::geom_textbox(data = filter(min_max_gps, dose == 2),
aes(x = case_when(dose < 1.5 ~ dose + 0.05, TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1, TRUE ~ len * 0.9),
label = paste0("**<span style='font-family:Enriqueta'>", guinea_pig_name,"</span>** - ", len, " mm"),
hjust = case_when(dose < 1.5 ~ 0,TRUE ~ 1),
halign = case_when(dose < 1.5 ~ 0, TRUE ~ 1)),
family = "Cabin", size = 4, fill = NA, box.colour = NA) +
geom_curve(data = filter(min_max_gps,
dose == 2 &
min_or_max == "max"),
aes(x = case_when(dose < 1.5 ~ dose + 0.05, TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1, TRUE ~ len * 0.9),
xend = case_when(dose < 1.5 ~ dose + 0.02, TRUE ~ dose - 0.02),
yend = case_when(min_or_max == "max"~ len + 0.5,TRUE ~ len - 0.5)),
curvature = -0.1,
arrow = arrow(length = unit(0.1, "cm")),
alpha = 0.5) +
geom_curve(data = filter(min_max_gps,
dose == 2 &
min_or_max == "min"),
aes(x = case_when(dose < 1.5 ~ dose + 0.05, TRUE ~ dose - 0.05),
y = case_when(min_or_max == "max"~ len * 1.1, TRUE ~ len * 0.9),
xend = case_when(dose < 1.5 ~ dose + 0.02, TRUE ~ dose - 0.02),
yend = case_when(min_or_max == "max"~ len + 0.5, TRUE ~ len - 0.5)),
curvature = 0.1,
arrow = arrow(length = unit(0.1, "cm")),
alpha = 0.5) +
scale_colour_manual(values = vit_c_palette)Curved arrows
To avoid it getting too unwieldy, add the curvatures to your data and iterate.
labelled_plot # plot with everything but the arrows
for(curv in unique(my_data$curvature)) {
filtered_data <- filter(my_data,
curvature == curv)
labelled_plot <- labelled_plot +
annotate(geom = "curve",
x = filtered_data$label_x,
y = filtered_data$label_y,
xend = filtered_data$arrow_end_x,
yend = filtered_data$arrow_end_y,
size = 0.3,
colour = case_when(filtered_data$species == "Adelie" ~ penguin_palette$Adelie,
filtered_data$species == "Chinstrap" ~ penguin_palette$Chinstrap,
filtered_data$species == "Gentoo" ~ penguin_palette$Gentoo),
curvature = curv,
arrow = arrow(length = unit(1.5, "mm")))
}
labelled_plot # plot with as many different curvatures as you like!Curved arrows
To avoid it getting too unwieldy, add the curvatures to your data and iterate.
labelled_plot # plot with everything but the arrows
for(curv in unique(my_data$curvature)) {
filtered_data <- filter(my_data,
curvature == curv)
labelled_plot <- labelled_plot +
annotate(geom = "curve",
x = filtered_data$label_x,
y = filtered_data$label_y,
xend = filtered_data$arrow_end_x,
yend = filtered_data$arrow_end_y,
size = 0.3,
colour = case_when(filtered_data$species == "Adelie" ~ penguin_palette$Adelie,
filtered_data$species == "Chinstrap" ~ penguin_palette$Chinstrap,
filtered_data$species == "Gentoo" ~ penguin_palette$Gentoo),
curvature = curv,
arrow = arrow(length = unit(1.5, "mm")))
}
labelled_plot # plot with as many different curvatures as you like!Find out more: cararthompson.com/talks/user2022
Curved arrows
Over to you!
hello@cararthompson.com
Tw/Li: @cararthompson