Beautifully reproducible dataviz
(mis)adventures in creating data-to-viz pipelines
Cara R Thompson | Building Stories with Data LTD
RPySOC 2025
Hello! 👋
👩 Cara Thompson
👩💻 Love for patterns in music & language, and a fascination with the human brain |>
Psychology PhD |>
Analysis of postgraduate medical examinations |>
Data Visualisation Consultant
💙 Helping others maximise the impact of their expertise
🛠️ Bespoke dataviz packages
Find out more: cararthompson.com/about
I ❤️ parameterising dataviz
I ❤️ parameterising dataviz
The why
A silly scenario
“We are welcoming a new group of penguins to the London zoo. We want to check their beak lengths, because our zoo keepers are nervous about penguins with long beaks. Can you help us visualise the beak lengths?”
- They’re coming from Edinburgh zoo, so we know the beak lengths in that group
- No idea how many, probably 50ish?
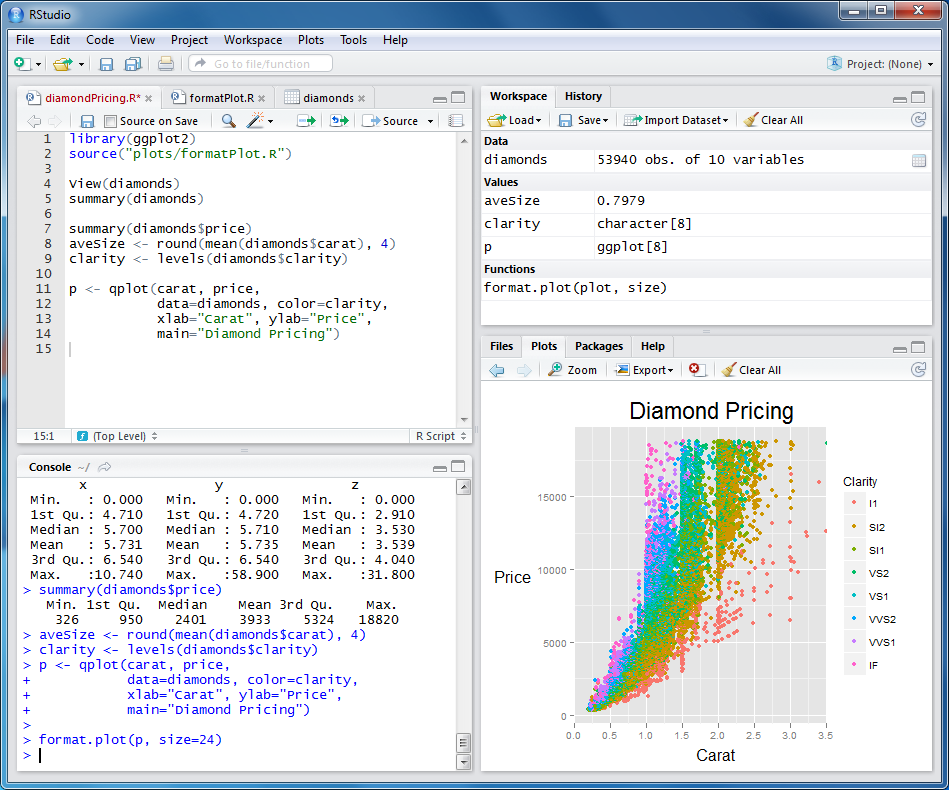
Look what I built!
# Libraries ----
library(ggplot2)
# Data wrangling ----
all_penguins <- palmerpenguins::penguins_raw |>
janitor::clean_names() |>
dplyr::filter(!is.na(culmen_length_mm)) |>
dplyr::mutate(species = gsub("(.)( )(.*)", "\\1", species))
set.seed(2134)
penguin_df <- all_penguins |>
dplyr::sample_n(50)
# Build a theme ----
theme_penguins <- function(
base_text_size = 20,
base_font = "Work Sans",
title_font = "Poppins"
) {
theme_minimal(base_size = base_text_size) +
theme(
text = element_text(family = base_font),
axis.text = element_text(colour = "#495058"),
legend.position = "none",
axis.title = element_blank(),
plot.title = element_text(
family = title_font,
face = "bold",
hjust = 0.5,
size = rel(1.5)
),
panel.grid = element_line(colour = "#FFFFFF"),
plot.background = element_rect(fill = "#F1F1F9", colour = "#F1F1F9"),
plot.margin = margin_auto(base_text_size * 2)
)
}
# Set up colours ----
penguin_colours <- c("#F49F03", "#F4B9C4", "#11541F")
# Create dataviz function ----
make_beak_plot <- function(df = penguin_df, colours = penguin_colours) {
beak_means_df <- df |>
dplyr::group_by(species) |>
dplyr::summarise(mean_length = mean(culmen_length_mm, na.rm = TRUE))
beak_range_df <- df |>
dplyr::filter(
culmen_length_mm == max(culmen_length_mm, na.rm = TRUE) |
culmen_length_mm == min(culmen_length_mm, na.rm = TRUE)
)
interactive_plot <- df |>
ggplot(aes(x = culmen_length_mm, y = species)) +
geom_vline(
data = beak_range_df,
aes(xintercept = culmen_length_mm),
linetype = 3,
colour = "#1A242F"
) +
geom_segment(
data = beak_means_df,
aes(x = mean_length, xend = mean_length, y = -Inf, yend = species),
linetype = 3
) +
ggiraph::geom_jitter_interactive(
aes(
x = culmen_length_mm,
y = species,
fill = species,
tooltip = paste0("<b>", individual_id, "</b> from ", island)
),
shape = 21,
width = 0,
size = 8,
height = 0.15,
colour = "#1A242F",
stroke = 0.5,
alpha = 0.9
) +
ggtext::geom_textbox(
data = beak_range_df,
aes(
y = max(species),
label = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~
paste0("🞀 ", culmen_length_mm, "mm"),
TRUE ~ paste0(culmen_length_mm, "mm", " 🞂")
),
hjust = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~ 0,
TRUE ~ 1
),
halign = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~ 0,
TRUE ~ 1
)
),
family = "Work Sans",
colour = "#1A242F",
fontface = "bold",
fill = NA,
size = 8,
box.padding = unit(0, "pt"),
box.colour = NA,
nudge_y = 0.33
) +
ggtext::geom_textbox(
data = beak_means_df,
aes(
x = mean_length,
y = species,
label = paste0(
species,
" mean<br>**",
janitor::round_half_up(mean_length),
"mm**"
),
hjust = dplyr::case_when(mean_length > 45 ~ 1, .default = 0),
halign = dplyr::case_when(mean_length > 45 ~ 1, .default = 0)
),
nudge_y = -0.3,
box.colour = NA,
size = 6,
family = "Work Sans",
colour = "#1A242F",
fill = NA
) +
labs(title = "Beak lengths by species") +
scale_fill_manual(values = colours) +
scale_x_continuous(label = function(x) paste0(x, "mm")) +
theme_penguins() +
theme(axis.text.y = element_blank())
ggiraph::girafe(
ggobj = interactive_plot,
options = list(ggiraph::opts_tooltip(
css = "background-color:#1A242F;color:#F4F5F6;padding:7.5px;letter-spacing:0.025em;line-height:1.3;border-radius:5px;font-family:Work Sans;"
)),
height_svg = 9
)
}
# Run function ----
make_beak_plot()“Could you just…?”
“Could you just…?”
Escaping from the copy-paste nightmare
- 🖼️ The theme
- 🎨 The colours
- 🛠️ Turn the code into a function
- 📦 Package everything up
- 🖼️ Reuse across all graphs
- 🎨 Reuse across all graphs
- 🛠️ No copy-pasting
- 📦 Dependencies + version control
RMedicine Workshop: Visualise, Optimise, Parameterise!
“Et finalement… la dataviz!”
“It is possible to generate R code that you can paste into your script to consistently generate the same look.”
- Bruno Rodrigues, Building Reproducible Analytical Pipelines with R, p. 423
“Can we use the new ggplot2 release…?”
The traps
Axis limits
“We need all the plots to be easily comparable”
Axis limits
“We need all the plots to be easily comparable”
make_beak_plot <- function(
df = penguin_df,
colours = penguin_colours
) {
beak_means_df <- df |>
dplyr::group_by(species) |>
dplyr::summarise(mean_length = mean(culmen_length_mm, na.rm = TRUE))
beak_range_df <- df |>
dplyr::filter(
culmen_length_mm == max(culmen_length_mm, na.rm = TRUE) |
culmen_length_mm == min(culmen_length_mm, na.rm = TRUE)
)
interactive_plot <- df |>
ggplot(aes(x = culmen_length_mm, y = species)) +
geom_vline(
data = beak_range_df,
aes(xintercept = culmen_length_mm),
linetype = 3,
colour = "#1A242F"
) +
geom_segment(
data = beak_means_df,
aes(x = mean_length, xend = mean_length, y = -Inf, yend = species),
linetype = 3
) +
ggiraph::geom_jitter_interactive(
aes(
x = culmen_length_mm,
y = species,
fill = species,
tooltip = paste0("<b>", individual_id, "</b> from ", island)
),
shape = 21,
width = 0,
size = 8,
height = 0.15,
colour = "#1A242F",
stroke = 0.5,
alpha = 0.9
) +
ggtext::geom_textbox(
data = beak_range_df,
aes(
y = max(species),
label = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~
paste0("🞀 ", culmen_length_mm, "mm"),
TRUE ~ paste0(culmen_length_mm, "mm", " 🞂")
),
hjust = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~ 0,
TRUE ~ 1
),
halign = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~ 0,
TRUE ~ 1
)
),
family = "Work Sans",
colour = "#1A242F",
fontface = "bold",
fill = NA,
size = 8,
box.padding = unit(0, "pt"),
box.colour = NA,
nudge_y = 0.33
) +
ggtext::geom_textbox(
data = beak_means_df,
aes(
x = mean_length,
y = species,
label = paste0(
species,
" mean<br>**",
janitor::round_half_up(mean_length),
"mm**"
),
hjust = dplyr::case_when(mean_length > 45 ~ 1, .default = 0),
halign = dplyr::case_when(mean_length > 45 ~ 1, .default = 0)
),
nudge_y = -0.3,
box.colour = NA,
size = 6,
family = "Work Sans",
colour = "#1A242F",
fill = NA
) +
labs(title = "Beak lengths by species") +
scale_fill_manual(values = colours) +
scale_x_continuous(
label = function(x) paste0(x, "mm"),
limits = c(32, 60)
) +
theme_penguins() +
theme(axis.text.y = element_blank())
ggiraph::girafe(
ggobj = interactive_plot,
options = list(ggiraph::opts_tooltip(
css = "background-color:#1A242F;color:#F4F5F6;padding:7.5px;letter-spacing:0.025em;line-height:1.3;border-radius:5px;font-family:Work Sans;"
)),
height_svg = 9
)
}Axis limits
“We need all the plots to be easily comparable”
What’s going on with those labels?
Quick bug fix!
What’s going on with those labels?
Quick bug fix!
make_beak_plot <- function(
df = penguin_df,
colours = penguin_colours
) {
beak_means_df <- df |>
dplyr::group_by(species) |>
dplyr::summarise(mean_length = mean(culmen_length_mm, na.rm = TRUE))
beak_range_df <- df |>
dplyr::filter(
culmen_length_mm == max(culmen_length_mm, na.rm = TRUE) |
culmen_length_mm == min(culmen_length_mm, na.rm = TRUE)
)
interactive_plot <- df |>
ggplot(aes(x = culmen_length_mm, y = species)) +
geom_vline(
data = beak_range_df,
aes(xintercept = culmen_length_mm),
linetype = 3,
colour = "#1A242F"
) +
geom_segment(
data = beak_means_df,
aes(x = mean_length, xend = mean_length, y = -Inf, yend = species),
linetype = 3
) +
ggiraph::geom_jitter_interactive(
aes(
x = culmen_length_mm,
y = species,
fill = species,
tooltip = paste0("<b>", individual_id, "</b> from ", island)
),
shape = 21,
width = 0,
size = 8,
height = 0.15,
colour = "#1A242F",
stroke = 0.5,
alpha = 0.9
) +
ggtext::geom_textbox(
data = beak_range_df,
aes(
# Used to be max(species), from the beak_range_df
y = max(df$species),
label = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~
paste0("🞀 ", culmen_length_mm, "mm"),
TRUE ~ paste0(culmen_length_mm, "mm", " 🞂")
),
hjust = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~ 0,
TRUE ~ 1
),
halign = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~ 0,
TRUE ~ 1
)
),
family = "Work Sans",
colour = "#1A242F",
fontface = "bold",
fill = NA,
size = 8,
box.padding = unit(0, "pt"),
box.colour = NA,
nudge_y = 0.33
) +
ggtext::geom_textbox(
data = beak_means_df,
aes(
x = mean_length,
y = species,
label = paste0(
species,
" mean<br>**",
janitor::round_half_up(mean_length),
"mm**"
),
hjust = dplyr::case_when(mean_length > 45 ~ 1, .default = 0),
halign = dplyr::case_when(mean_length > 45 ~ 1, .default = 0)
),
nudge_y = -0.3,
box.colour = NA,
size = 6,
family = "Work Sans",
colour = "#1A242F",
fill = NA
) +
labs(title = "Beak lengths by species") +
scale_fill_manual(values = colours) +
scale_x_continuous(
label = function(x) paste0(x, "mm"),
limits = c(32, 60)
) +
theme_penguins() +
theme(axis.text.y = element_blank())
ggiraph::girafe(
ggobj = interactive_plot,
options = list(ggiraph::opts_tooltip(
css = "background-color:#1A242F;color:#F4F5F6;padding:7.5px;letter-spacing:0.025em;line-height:1.3;border-radius:5px;font-family:Work Sans;"
)),
height_svg = 9
)
}What’s going on with those labels?
Quick bug fix!
Colours
“The penguin species should in overall length order”
Colours
“The penguin species should in overall length order”
Colours
“The penguin species should in overall length order”
Why are the dots dancing around?
Jitter in geom_jitter or in the data?
Why are the dots dancing around?
Jitter in geom_jitter or in the data?
make_beak_plot <- function(
df = penguin_df,
colours = penguin_colours
) {
beak_means_df <- df |>
dplyr::group_by(species) |>
dplyr::summarise(mean_length = mean(culmen_length_mm, na.rm = TRUE))
beak_range_df <- df |>
dplyr::filter(
culmen_length_mm == max(culmen_length_mm, na.rm = TRUE) |
culmen_length_mm == min(culmen_length_mm, na.rm = TRUE)
)
interactive_plot <- df |>
ggplot(aes(x = culmen_length_mm, y = as.numeric(species))) +
geom_vline(
data = beak_range_df,
aes(xintercept = culmen_length_mm),
linetype = 3,
colour = "#1A242F"
) +
geom_segment(
data = beak_means_df,
aes(
x = mean_length,
xend = mean_length,
y = -Inf,
yend = as.numeric(species)
),
linetype = 3
) +
ggiraph::geom_point_interactive(
aes(
x = culmen_length_mm,
y = as.numeric(species) + jitter_y,
fill = species,
tooltip = paste0("<b>", individual_id, "</b> from ", island)
),
shape = 21,
width = 0,
size = 8,
colour = "#1A242F",
stroke = 0.5,
alpha = 0.9
) +
ggtext::geom_textbox(
data = beak_range_df,
aes(
# Used to be max(species), from the beak_range_df
y = max(as.numeric(df$species)),
label = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~
paste0("🞀 ", culmen_length_mm, "mm"),
TRUE ~ paste0(culmen_length_mm, "mm", " 🞂")
),
hjust = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~ 0,
TRUE ~ 1
),
halign = dplyr::case_when(
culmen_length_mm == min(culmen_length_mm) ~ 0,
TRUE ~ 1
)
),
family = "Work Sans",
colour = "#1A242F",
fontface = "bold",
fill = NA,
size = 8,
box.padding = unit(0, "pt"),
box.colour = NA,
nudge_y = 0.33
) +
ggtext::geom_textbox(
data = beak_means_df,
aes(
x = mean_length,
y = as.numeric(species),
label = paste0(
species,
" mean<br>**",
janitor::round_half_up(mean_length),
"mm**"
),
hjust = dplyr::case_when(mean_length > 45 ~ 1, .default = 0),
halign = dplyr::case_when(mean_length > 45 ~ 1, .default = 0)
),
nudge_y = -0.3,
box.colour = NA,
size = 6,
family = "Work Sans",
colour = "#1A242F",
fill = NA
) +
labs(title = "Beak lengths by species") +
scale_fill_manual(values = colours) +
scale_y_continuous(breaks = c(1, 2, 3)) +
scale_x_continuous(
label = function(x) paste0(x, "mm"),
limits = c(32, 60)
) +
theme_penguins() +
theme(axis.text.y = element_blank(),
panel.grid.minor.y = element_blank())
ggiraph::girafe(
ggobj = interactive_plot,
options = list(ggiraph::opts_tooltip(
css = "background-color:#1A242F;color:#F4F5F6;padding:7.5px;letter-spacing:0.025em;line-height:1.3;border-radius:5px;font-family:Work Sans;"
)),
height_svg = 9
)
}Why are the dots dancing around?
Jitter in geom_jitter or in the data?
Why are the dots dancing around?
Jitter in geom_jitter or in the data?
Why are the dots dancing around?
Jitter in geom_jitter or in the data?
Hello, real world!
(Very) different data
I thought you said “50ish?”
Different interactions
Different interactions
“The tooltips don’t work for me”
Different tools
RStudio vs Positron vs VS Code …
Fonts not installed
“Wait, it doesn’t look the same on my computer…”
Blog post: Getting fonts to work in R
Different… everything?
“I’m getting an error message…”
Artwork by Allison Horst
New {ggplot2} release!
Testing, testing, 1-2-3

Sensible tests
- Predictable / eyeball-able (does the visualisation match the data?)
- Edge cases (does our
dataviz_function()work in odd circumstances?) - All the traps we fixed earlier
- Any future “Oh, that’s interesting…” moments
Unknown unknowns
Create a reference ggplot object for each permutation, and use testthat::expect_equal() or waldo::compare() to check for differences?
- Jitter not picked up
- Font issue not picked up
- Interpretation of the ggplot code
Unknown unknowns
Quick demo
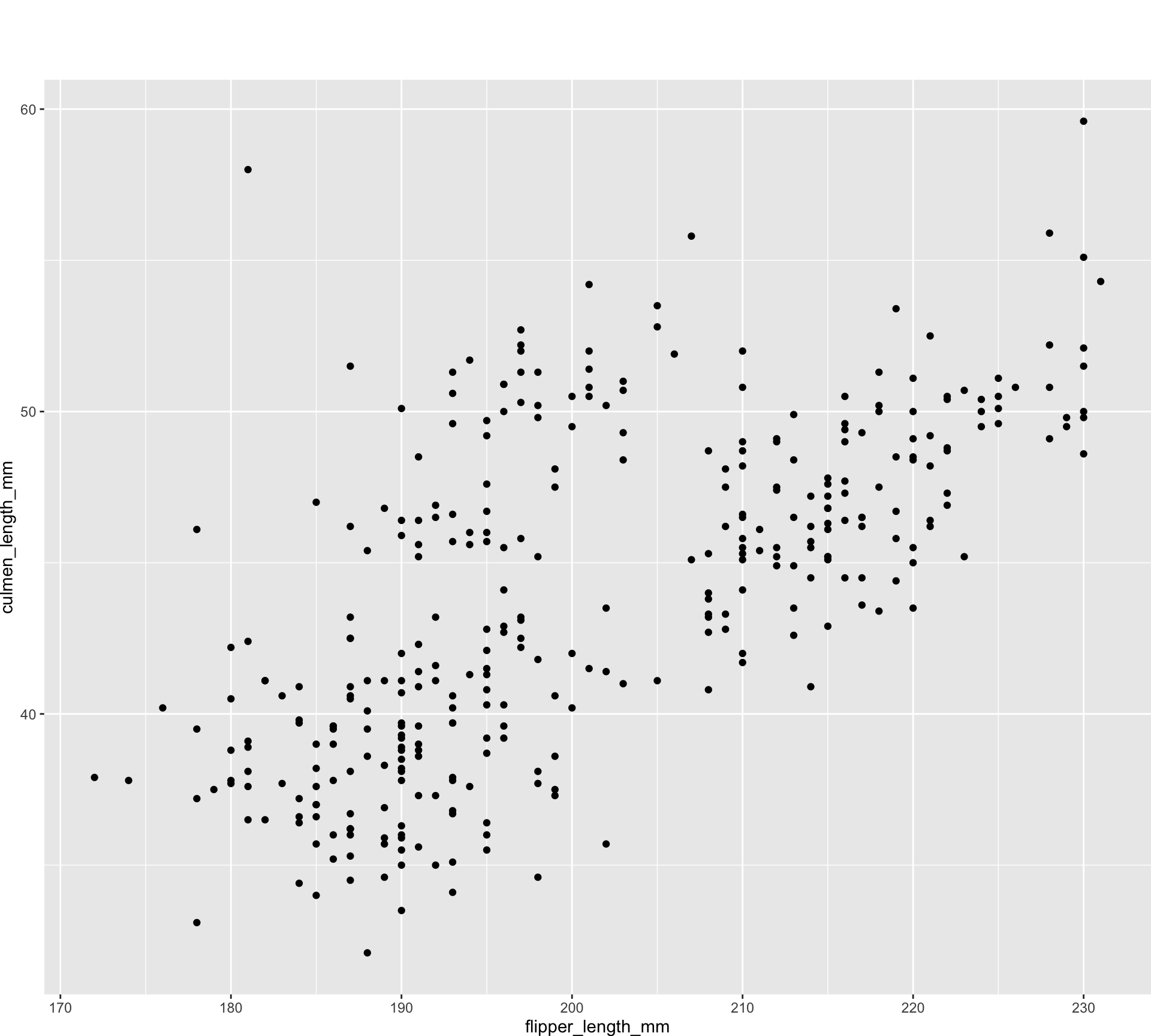
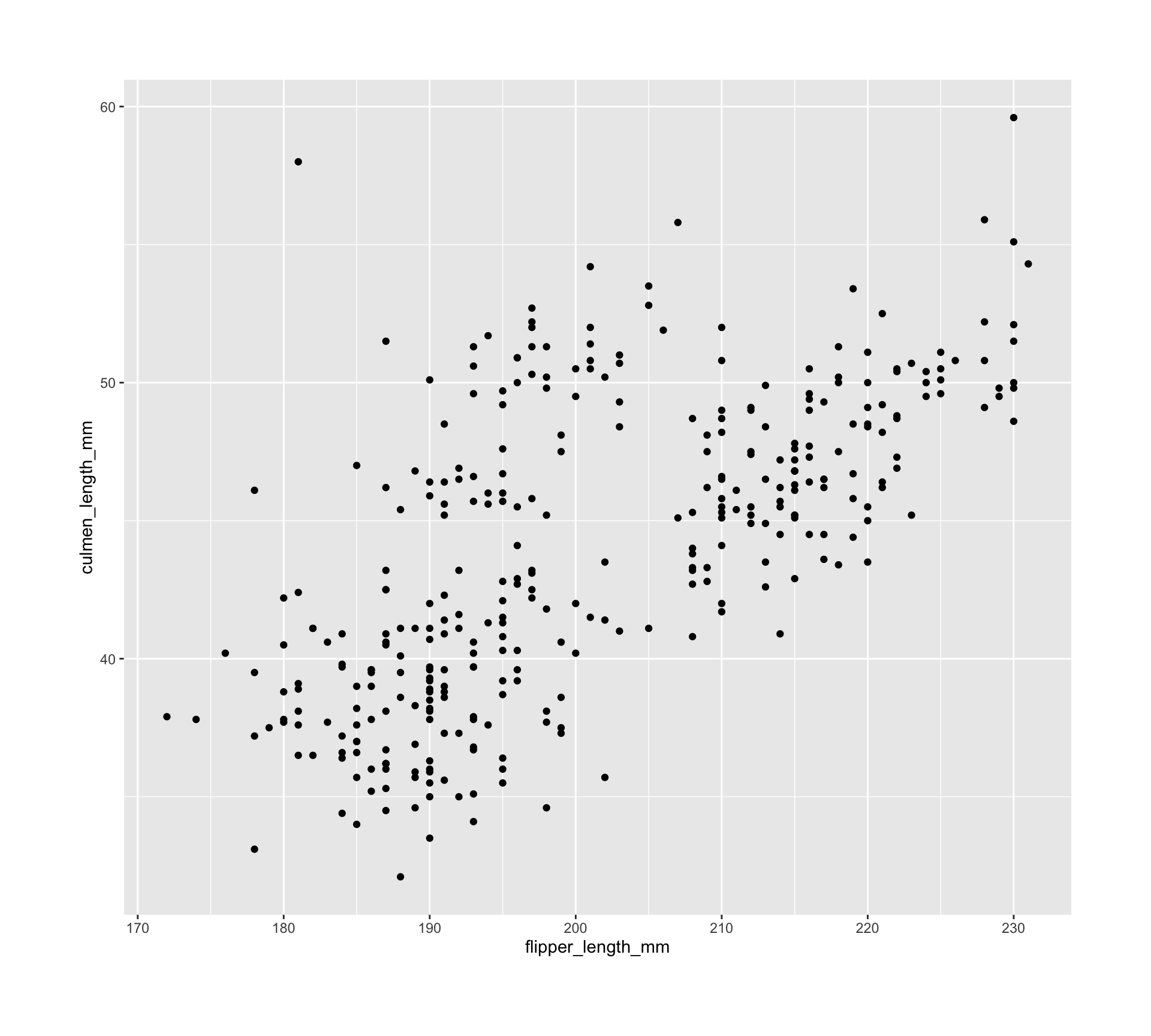
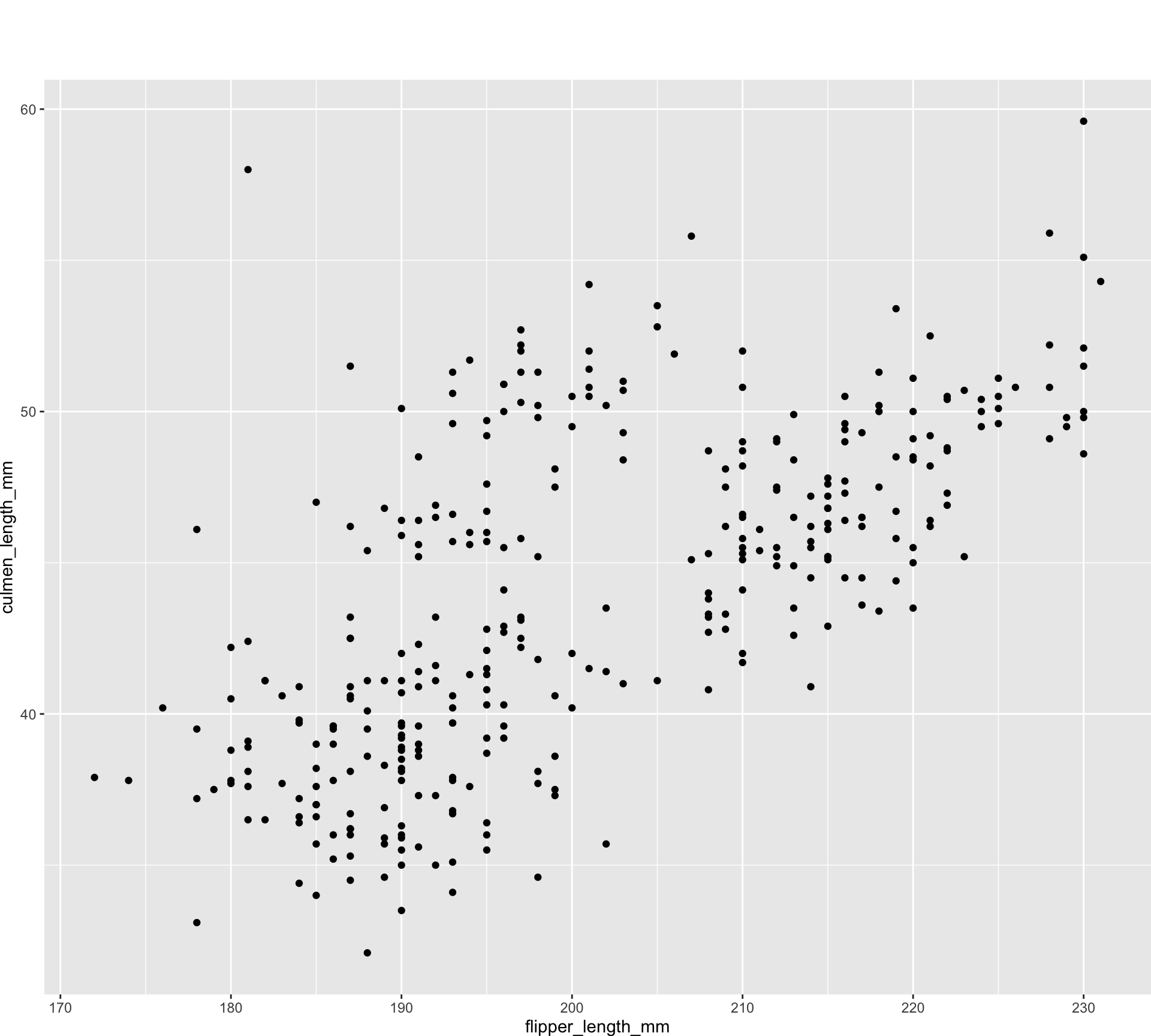
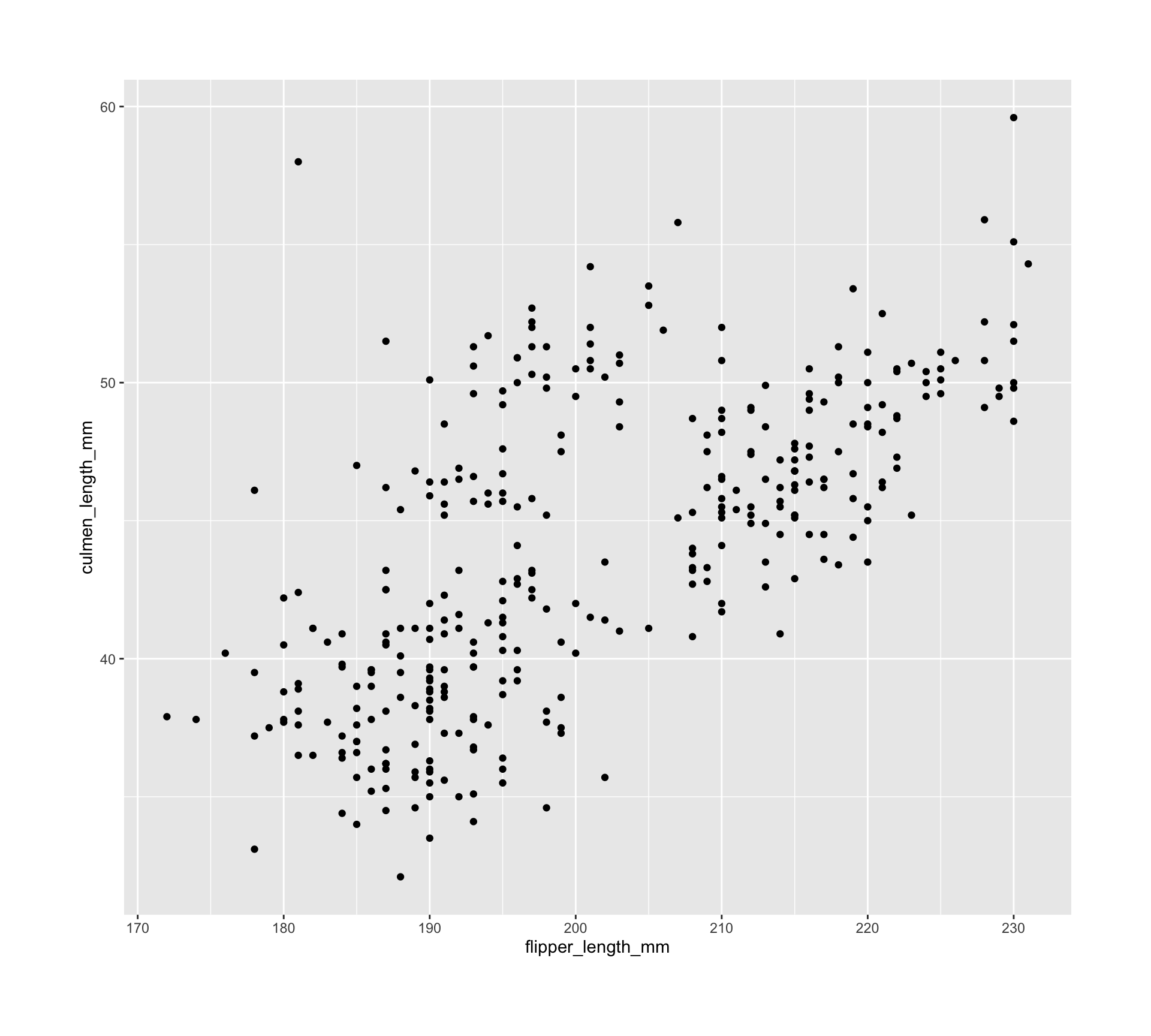
old_margin_trick <- ggplot(all_penguins) +
geom_point(aes(x = flipper_length_mm, y = culmen_length_mm)) +
theme(plot.margin = margin(rep(50, 4)))
new_margin_way <- ggplot(all_penguins) +
geom_point(aes(x = flipper_length_mm, y = culmen_length_mm)) +
theme(plot.margin = margin(50, 50, 50, 50))
new_margin_auto <- ggplot(all_penguins) +
geom_point(aes(x = flipper_length_mm, y = culmen_length_mm)) +
theme(plot.margin = margin_auto(50))Unknown unknowns
Unknown unknowns
Unknown unknowns
waldo::compare(old_margin_trick, new_margin_auto)
`old@theme$plot.margin`: "50points" "0points" "0points" "0points"
`new@theme$plot.margin`: "50points" "50points" "50points" "50points"waldo::compare(new_margin_way, new_margin_auto)
`old@layers$geom_point` is length 14
`new@layers$geom_point` is length 17
names(old@layers$geom_point) | names(new@layers$geom_point)
[1] "aes_params" | "aes_params" [1]
- "computed_geom_params" [2]
- "computed_mapping" [3]
- "computed_stat_params" [4]
[2] "constructor" | "constructor" [5]
[3] "data" | "data" [6]
[4] "geom" | "geom" [7]
`old@layers$geom_point$computed_geom_params` is absent
`new@layers$geom_point$computed_geom_params` is a list
`old@layers$geom_point$computed_mapping` is absent
`new@layers$geom_point$computed_mapping` is an S7 object of class <ggplot2::mapping>
`old@layers$geom_point$computed_stat_params` is absent
`new@layers$geom_point$computed_stat_params` is a listUnknown unknowns
An imperfect solution
- Make a reference plot
- Save as PNG
- ⚠️ Update code ⚠️
- Make a new plot
- Save as PNG
- Read them both back in using
png::readPNG() - Compare using
testhat::expect_equal()
{vdiffr} - a much better solution!
Similar to {testthat}:
- within a package
- set up tests
- generate dopplegangers when run tests for the first time
- change code, run tests again, use viewer to see the difference!
{vdiffr} - quick demo
mypackage/tests/testthat/test_ggplot_outputs.R
set.seed(2025) # IMPORTANT!
all_penguins <- palmerpenguins::penguins_raw |>
janitor::clean_names() |>
dplyr::filter(!is.na(culmen_length_mm)) |>
dplyr::mutate(species = gsub("(.)( )(.*)", "\\1", species)) |>
dplyr::mutate(
species = factor(
species,
levels = c("Adelie", "Gentoo", "Chinstrap")
)
) |>
dplyr::rowwise() |>
dplyr::mutate(jitter_y = sample(runif(100, -0.1, 0.1), 1)) |>
dplyr::ungroup()
penguin_sample_1 <- all_penguins |>
dplyr::filter(species == "Gentoo")
penguin_sample_2 <- all_penguins |>
dplyr::filter(culmen_length_mm < 45)
set.seed(1234)
penguin_sample_3 <- all_penguins |>
dplyr::sample_n(50)
# ...
describe("dataviz function", {
it("The function works with all the penguins", {
vdiffr::expect_doppelganger("all penguins", make_beak_plot(all_penguins))
})
})
describe("dataviz function", {
it("The function works with only Gentoo", {
vdiffr::expect_doppelganger("sample 1 Gentoo", make_beak_plot(penguin_sample_1))
})
})
describe("dataviz function", {
it("The function works short beaks", {
vdiffr::expect_doppelganger("sample 2 short beaks", make_beak_plot(penguin_sample_2))
})
})
describe("dataviz function", {
it("The function works with sample of 50", {
vdiffr::expect_doppelganger("sample 3 seed12334 50", make_beak_plot(penguin_sample_3))
})
})
# ...{vdiffr} - quick demo
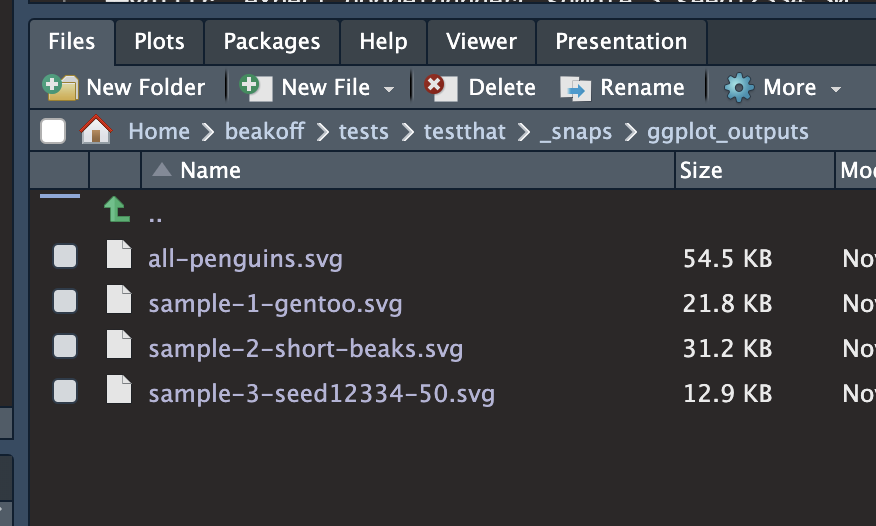
mypackage/tests/testthat/_snaps/ggplot_outputs
{vdiffr} - quick demo
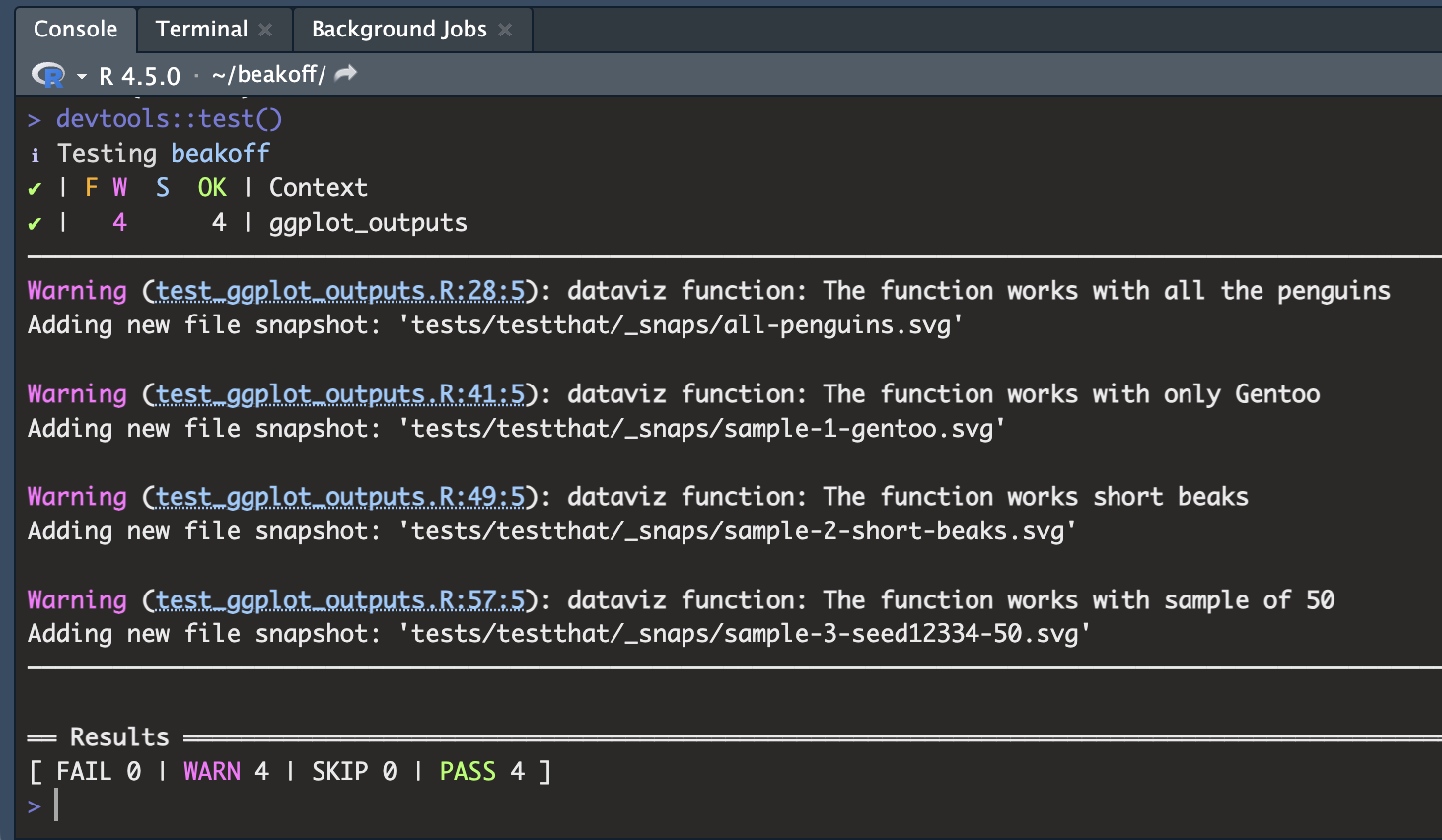
It tells you what it’s doing 😊
{vdiffr} - quick demo
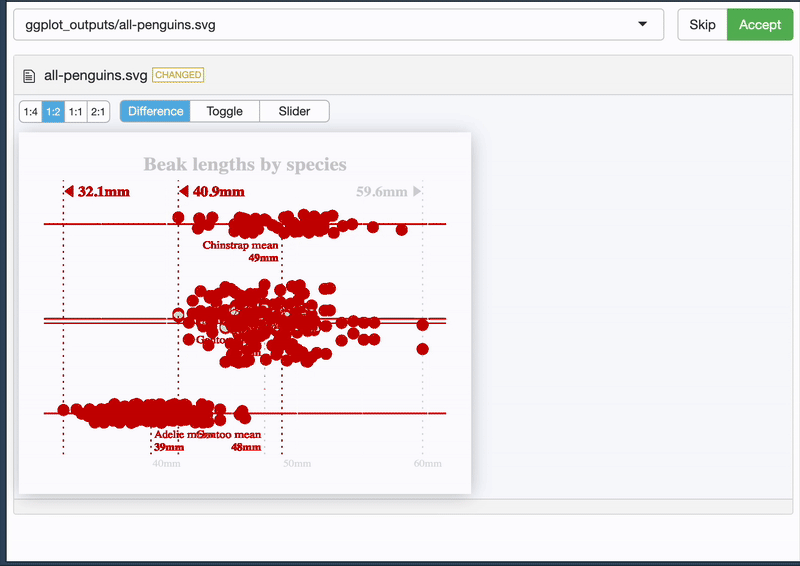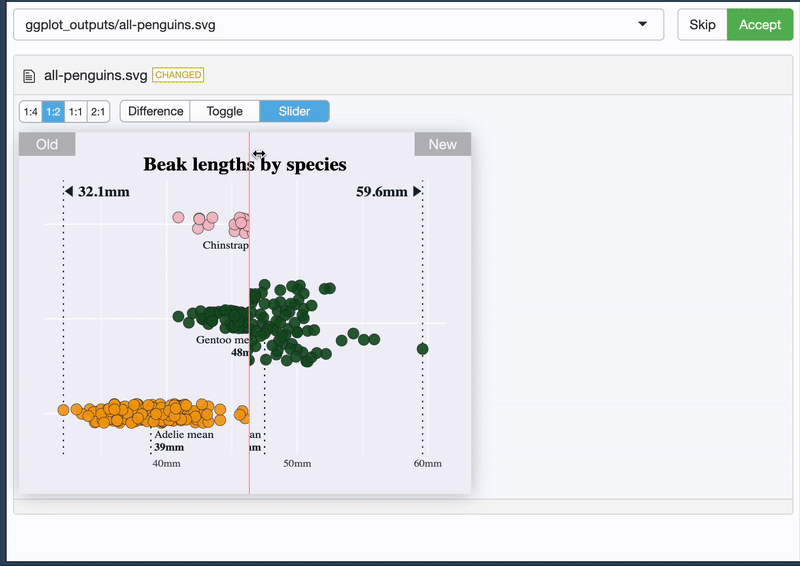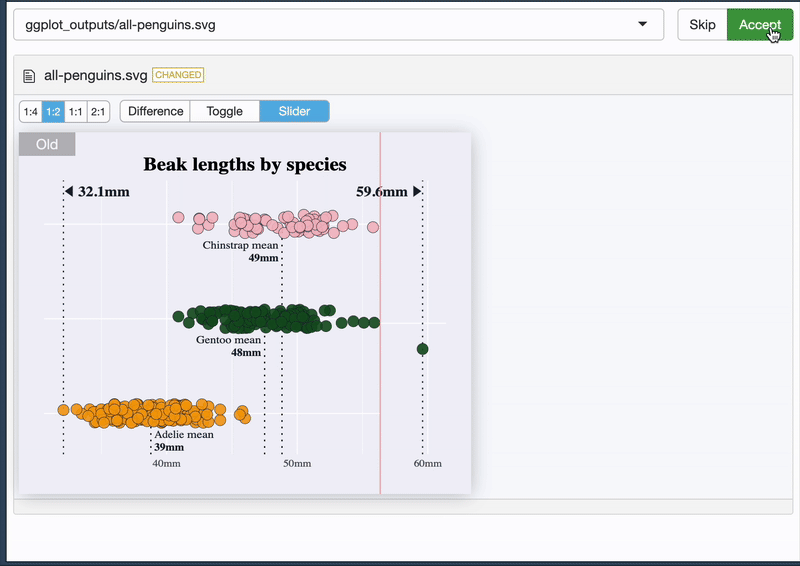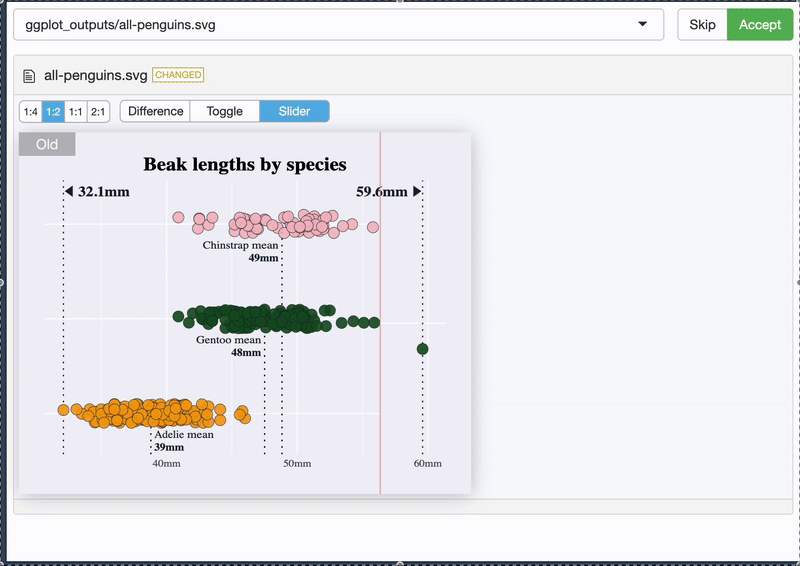
And it allows you to see the difference 🥳
{vdiffr} - quick demo
And it allows you to see the difference 🥳
{vdiffr} - quick demo
And it allows you to see the difference 🥳
- Fonts?
- Interactivity?
- …
(Thank you to Lesley Duff for getting me this far!)
Bye bye, Boy-zer
Bye bye, Boy-zer
Let’s chat!
cararthompson.com/talks